

| 機種名 | 台数 | 性能 | ||
|---|---|---|---|---|
| 平均解読長 | データ量 | ラン時間 | ||
| NovaSeq 6000(Illumina) | 2 | 150bp | ~6Tb | 45時間 |
| NextSeq 2000(Illumina) | 1 | 150bp | 〜540Gb | 44時間 |
| AVITI / AVITI 24 (Element Biosciences) | 1/1 | 150bp | ~255Gb | 52時間 |
| Sequel Ⅱe(Pacific Biosciences) | 1 | ~20kb | ~30Gb | 30時間 |
| Revio(Pacific Biosciences) | 1 | ~20kb | ~90Gb | 30時間 |
| PromethION(Oxford Nanopore) | 1 | ライブラリに依存 | ~100Gb | ~72時間 |
| GridION(Oxford Nanopore) | 1 | ライブラリに依存 | ~10Gb | ~72時間 |







 | センター長 黒川 顕/教授
|  | シーケンシング部門長 豊田 敦/特任教授
| ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
 | データ解析部門長 森 宙史/准教授
|  | シーケンシング部門 後藤 恭宏/准教授
| ||||||||
 | データ解析部門 小原 雄治/特任教授
|  | データ解析部門 藤山 秋佐夫/特命教授
| ||||||||
 | データ解析部門 野口 英樹/特任教授
|  | データ解析部門 馬場 知哉/特任准教授
| ||||||||
 | データ解析部門 小杉 俊一/特任准教授
|  | データ解析部門 東 光一/助教
| ||||||||
 | データ解析部門 黒川 真臣/特任研究員
|

NGSが産出する膨大な情報から新規知識発見ができるようバイオインフォマティクス技術を駆使した解析を実施します。研究コンサルティングを通して、シーケンシング技術と一体化し、課題に適した解析技術を提供します。また、お手持ちのデータに対する情報解析も受け付けています。

| 機種名 | 台数 | 性能 | ||
|---|---|---|---|---|
| 平均解読長 | データ量 | ラン時間 | ||
| NovaSeq 6000(Illumina) | 2 | 150bp | ~6Tb | 45時間 |
| NextSeq 2000(Illumina) | 1 | 150bp | 〜540Gb | 44時間 |
| AVITI / AVITI 24 (Element Biosciences) | 1/1 | 150bp | ~255Gb | 52時間 |
| Sequel Ⅱe(Pacific Biosciences) | 1 | ~20kb | ~30Gb | 30時間 |
| Revio(Pacific Biosciences) | 1 | ~20kb | ~90Gb | 30時間 |
| PromethION(Oxford Nanopore) | 1 | ライブラリに依存 | ~100Gb | ~72時間 |
| GridION(Oxford Nanopore) | 1 | ライブラリに依存 | ~10Gb | ~72時間 |






 | センター長 黒川 顕/教授
|  | シーケンシング部門長 豊田 敦/特任教授
| ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
 | データ解析部門長 森 宙史/准教授
|  | シーケンシング部門 後藤 恭宏/准教授
| ||||||||
 | データ解析部門 小原 雄治/特任教授
|  | データ解析部門 藤山 秋佐夫/特命教授
| ||||||||
 | データ解析部門 野口 英樹/特任教授
|  | データ解析部門 馬場 知哉/特任准教授
| ||||||||
 | データ解析部門 小杉 俊一/特任准教授
|  | データ解析部門 東 光一/助教
| ||||||||
 | データ解析部門 黒川 真臣/特任研究員
|
NGSが産出する膨大な情報から新規知識発見ができるようバイオインフォマティクス技術を駆使した解析を実施します。研究コンサルティングを通して、シーケンシング技術と一体化し、課題に適した解析技術を提供します。また、お手持ちのデータに対する情報解析も受け付けています。

●動物:ヒト、チンパンジー、ニホンザル、マウス、ラット、アマミトゲネズミ、スンクス、イヌ、クジラ、イルカ、ワラビー、ニワトリ、コウモリ、アフリカツメガエル、シーラカンス、ポリプテルス、シクリッド、メダカ、キンギョ、アオイガイ、ナメクジウオ、ホヤ、線虫、ショウジョウバエ、テントウムシ、アゲハ、カイコ、クワコ、プラナリア、クマムシなど
●植物:イネ、シロイヌナズナ、アサガオ、ナンヨウアブラギリ、トマト、イヌビワ、ヒメツリガネゴケ、ワサビ、トウガラシ、ハナビシソウ、甘草、サンゴ共生褐虫藻、緑藻類、紅藻類、接合藻類、微細藻類など
●微生物:分裂酵母、麹菌、病原真菌、ヒト常在菌、病原菌、光合成菌、納豆菌、極限環境細菌類、共生細菌類、難培養細菌、新型コロナウイルスなど
●メタゲノム:ヒト(腸内、皮膚、口腔)、マウス、昆虫、豚、海洋、河川、湖沼、土壌、温泉、活性汚泥、排水、農地/液肥など
Satoshi Ansai, Atsushi Toyoda, Kohta Yoshida, Jun Kitano. Repositioning of centromere‐associated repeats during karyotype evolution in Oryzias fishes. Molecular Ecology 33,e17222 (2024). doi:10.1111/mec.17222
Takeshi Fujino, Katsushi Yamaguchi, Toshiyuki T. Yokoyama, Toshiya Hamanaka, Yoritaka Harazono, Hiroaki Kamada, Wataru Kobayashi, Tokuko Ujino-Ihara, Kentaro Uchiyama, Asako Matsumoto, Ayako Izuno, Yoshihiko Tsumura, Atsushi Toyoda, Shuji Shigenobu, Yoshinari Moriguchi, Saneyoshi Ueno, Masahiro Kasahara. A chromosome-level genome assembly of a model conifer plant, the Japanese cedar, Cryptomeria japonica D. Don. BMC Genomics 25, (2024). doi:10.1186/s12864-024-10929-4
Takanori Matsumaru, Toshiki Iwamatsu, Kana Ishigami, Makoto Inai, Wataru Kanto, Ayumi Ishigaki, Atsushi Toyoda, Satoshi Shuto, Katsumi Maenaka, Shinichi Nakagawa, Hiroshi Maita. Identification of
Keishi Kubota, Mion Oishi, Eigo Taniguchi, Akiho Akazawa, Katsunori Matsui, Kazuyoshi Kitazaki, Atsushi Toyoda, Hidehiro Toh, Hiroaki Matsuhira, Yosuke Kuroda, Tomohiko Kubo. Mitochondrial phylogeny and distribution of cytoplasmic male sterility-associated genes in Beta vulgaris. PLOS ONE 19, e0308551(2024). doi:10.1371/journal.pone.0308551
Shunya Kaneko, Keita Miyoshi, Kotaro Tomuro, Makoto Terauchi, Ryoya Tanaka, Shu Kondo, Naoki Tani, Kei-Ichiro Ishiguro, Atsushi Toyoda, Azusa Kamikouchi, Hideki Noguchi, Shintaro Iwasaki, Kuniaki Saito. Mettl1-dependent m7G tRNA modification is essential for maintaining spermatogenesis and fertility in Drosophila melanogaster. Nature Communications 15, (2024). doi:10.1038/s41467-024-52389-0
Keisuke Noguchi, Hidefumi Suzuki, Ryota Abe, Keiko Horiuchi, Rena Onoguchi-Mizutani, Nobuyoshi Akimitsu, Shintaro Ogawa, Tomohiko Akiyama, Yoko Ike, Yoko Ino, Yayoi Kimura, Akihide Ryo, Hiroshi Doi, Fumiaki Tanaka, Yutaka Suzuki, Atsushi Toyoda, Yuki Yamaguchi, Hidehisa Takahashi. Multi-omics analysis using antibody-based in situ biotinylation technique suggests the mechanism of Cajal body formation. Cell Reports 43, 114734(2024). doi:10.1016/j.celrep.2024.114734
Jaeha Kim, Takumi Murakami, Atsushi Toyoda, Hiroshi Mori. Metagenome-assembled genome sequence of Spiroplasma phoeniceum, assembled from the hindgut of Locusta migratoria, a migration grasshopper species. Microbiology Resource Announcements 13, (2024). doi:10.1128/mra.00353-24
Genta Okude, Yo Y. Yamasaki, Atsushi Toyoda, Seiichi Mori, Jun Kitano. Genome-wide analysis of histone modifications can contribute to the identification of candidate cis-regulatory regions in the threespine stickleback fish. BMC Genomics 25, (2024). doi:10.1186/s12864-024-10602-w
Jung Lee, Toshiaki Fujimoto, Katsushi Yamaguchi, Shuji Shigenobu, Ken Sahara, Atsushi Toyoda, Toru Shimada. W chromosome sequences of two bombycid moths provide an insight into the origin of Fem. Molecular Ecology 33, (2024). doi:10.1111/mec.17434
Adilgazy Semeigazin, Shiori Iida, Katsuhiko Minami, Sachiko Tamura, Satoru Ide, Koichi Higashi, Atsushi Toyoda, Ken Kurokawa, Kazuhiro Maeshima. Behaviors of nucleosomes with mutant histone H4s in euchromatic domains of living human cells. Histochemistry and Cell Biology 162, 23-40(2024). doi:10.1007/s00418-024-02293-x
Kazuki Furugori, Hidefumi Suzuki, Ryota Abe, Keiko Horiuchi, Tomohiko Akiyama, Tomonori Hirose, Atsushi Toyoda, Hidehisa Takahashi. Chimera
Kanta K. Ochiai, Daiki Hanawa, Harumi A. Ogawa, Hiroyuki Tanaka, Kazuma Uesaka, Tomoya Edzuka, Maki Shirae‐Kurabayashi, Atsushi Toyoda, Takehiko Itoh, Gohta Goshima. Genome sequence and cell biological toolbox of the highly regenerative, coenocytic green feather alga Bryopsis. The Plant Journal 119, 1091-1111(2024). doi:10.1111/tpj.16764
Makiko Kosugi, Shuji Ohtani, Kojiro Hara, Atsushi Toyoda, Hiroyo Nishide, Shin-Ichiro Ozawa, Yuichiro Takahashi, Yasuhiro Kashino, Sakae Kudoh, Hiroyuki Koike, Jun Minagawa. Characterization of the far-red light absorbing light-harvesting chlorophyll a/b binding complex, a derivative of the distinctive Lhca gene family in green algae. Frontiers in Plant Science 15, (2024). doi:10.3389/fpls.2024.1409116
Junji Konuma, Tomochika Fujisawa, Tomoaki Nishiyama, Masahiro Kasahara, Tomoko F Shibata, Masafumi Nozawa, Shuji Shigenobu, Atsushi Toyoda, Mitsuyasu Hasebe, Teiji Sota. Odd-Paired is Involved in Morphological Divergence of Snail-Feeding Beetles. Molecular Biology and Evolution 41, (2024). doi:10.1093/molbev/msae110
Haruka Aoyama, Toshihiro Arae, Yui Yamashita, Atsushi Toyoda, Satoshi Naito, Naoyuki Sotta, Yukako Chiba. Impact of translational regulation on diel expression revealed by time‐series ribosome profiling in Arabidopsis. The Plant Journal 118, 1889-1906(2024). doi:10.1111/tpj.16716
Mika Uehara, Takashi Inoue, Sumitaka Hase, Erika Sasaki, Atsushi Toyoda, Yasubumi Sakakibara. Decoding host-microbiome interactions through co-expression network analysis within the non-human primate intestine. mSystems 9, (2024). doi:10.1128/msystems.01405-23
Issei Yahiro, Oga Sato, Sipra Mohapatra, Koki Mukai, Atsushi Toyoda, Takehiko Itoh, Michiya Matsuyama, Tapas Chakraborty, Kohei Ohta. SDF-1/CXCR4 signal is involved in the induction of Primordial Germ Cell migration in a model marine fish, Japanese anchovy (Engraulis japonicus). General and Comparative Endocrinology 351, 114476(2024). doi:10.1016/j.ygcen.2024.114476
Kiyoshi Yamazaki, Yoshihiro Ohmori, Hirokazu Takahashi, Atsushi Toyoda, Yutaka Sato, Mikio Nakazono, Toru Fujiwara. Transcriptome Analysis of Rice Root Tips Reveals Auxin, Gibberellin and Ethylene Signaling Underlying Nutritropism. Plant And Cell Physiology 65, 671-679(2024). doi:10.1093/pcp/pcae003
Yasuhiko Kato, Joel H. Nitta, Christelle Alexa Garcia Perez, Nikko Adhitama, Pijar Religia, Atsushi Toyoda, Wataru Iwasaki, Hajime Watanabe. Identification of gene isoforms and their switching events between male and female embryos of the parthenogenetic crustacean Daphnia magna. Scientific Reports 14, (2024). doi:10.1038/s41598-024-59774-1
Takaya Yamasaki, Akira Nishiyama, Nagomi Kurogi, Koutarou Nishimura, Shion Nishida, Daisuke Kurotaki, Tatsuma Ban, Jordan A. Ramilowski, Keiko Ozato, Atsushi Toyoda, Tomohiko Tamura. Physical and functional interaction among Irf8 enhancers during dendritic cell differentiation. Cell Reports 43, 114107(2024). doi:10.1016/j.celrep.2024.114107
Yoshihito Niimura, Bhim B Biswa, Takushi Kishida, Atsushi Toyoda, Kazumichi Fujiwara, Masato Ito, Kazushige Touhara, Miho Inoue-Murayama, Scott H Jenkins, Christopher Adenyo, Boniface B Kayang, Tsuyoshi Koide. Synchronized Expansion and Contraction of Olfactory, Vomeronasal, and Taste Receptor Gene Families in Hystricomorph Rodents. Molecular Biology and Evolution 41, (2024). doi:10.1093/molbev/msae071
Kentaro Fukuta, Dai-ichiro Kato, Juri Maeda, Atsuhiro Tsuruta, Hirobumi Suzuki, Yukio Nagano, Hisao Tsukamoto, Kazuki Niwa, Makoto Terauchi, Atsushi Toyoda, Asao Fujiyama, Hideki Noguchi. Genome assembly of Genji firefly (Nipponoluciola cruciata) reveals novel luciferase-like luminescent proteins without peroxisome targeting signal. DNA Research 31, (2024). doi:10.1093/dnares/dsae006
Kaoru Yamano, Akane Haseda, Keisuke Iwabuchi, Takayuki Osabe, Yuki Sudo, Babil Pachakkil, Keisuke Tanaka, Yutaka Suzuki, Atsushi Toyoda, Hideki Hirakawa, Yasuyuki Onodera. QTL analysis of femaleness in monoecious spinach and fine mapping of a major QTL using an updated version of chromosome-scale pseudomolecules. PLOS ONE 19, e0296675(2024). doi:10.1371/journal.pone.0296675
Issei Yahiro, Kyle Dominic Eguid Barnuevo, Oga Sato, Sipra Mohapatra, Atsushi Toyoda, Takehiko Itoh, Kaoru Ohno, Michiya Matsuyama, Tapas Chakraborty, Kohei Ohta. Modeling the SDF-1/CXCR4 protein using advanced artificial intelligence and antagonist screening for Japanese anchovy. Frontiers in Physiology 15, (2024). doi:10.3389/fphys.2024.1349119
Takahiko Kubo, Yoshiyuki Yamagata, Hiroaki Matsusaka, Atsushi Toyoda, Yutaka Sato, Toshihiro Kumamaru. MiRiQ Database: A Platform for In Silico Rice Mutant Screening. Plant And Cell Physiology 65, 169-174(2024). doi:10.1093/pcp/pcad134
Tomoya Ishii, Natsuki Tsuchida, Niarsi Merry Hemelda, Kirara Saito, Jiyuan Bao, Megumi Watanabe, Atsushi Toyoda, Takehiro Matsubara, Mayuko Sato, Kiminori Toyooka, Nobuaki Ishihama, Ken Shirasu, Hidenori Matsui, Kazuhiro Toyoda, Yuki Ichinose, Tetsuya Hayashi, Akira Kawaguchi, Yoshiteru Noutoshi. Rhizoviticin is an alphaproteobacterial tailocin that mediates biocontrol of grapevine crown gall disease. The ISME Journal 18, (2024). doi:10.1093/ismejo/wrad003
Luisa Matiz-Ceron, Miki Okuno, Takehiko Itoh, Ikuya Yoshida, Shusei Mizushima, Atsushi Toyoda, Takamichi Jogahara, Asato Kuroiwa. Loss of One X and the Y Chromosome Changes the Configuration of the X Inactivation Center in the Genus Tokudaia Cytogenetic and Genome Research 164, 23-32(2024). doi:10.1159/000539294
Miki Okuno, Yuta Mochimaru, Kentaro Matsuoka, Takahiro Yamabe, Luisa Matiz-Ceron, Takamichi Jogahara, Atsushi Toyoda, Asato Kuroiwa, Takehiko Itoh. Chromosomal-level assembly of Tokudaia osimensis, Tokudaia tokunoshimensis, and Tokudaia muenninki genomes. Scientific Data 10, (2023). doi:10.1038/s41597-023-02845-1
Muran Xiao, Shinji Kondo, Masaki Nomura, Shinichiro Kato, Koutarou Nishimura, Weijia Zang, Yifan Zhang, Tomohiro Akashi, Aaron Viny, Tsukasa Shigehiro, Tomokatsu Ikawa, Hiromi Yamazaki, Miki Fukumoto, Atsushi Tanaka, Yasutaka Hayashi, Yui Koike, Yumi Aoyama, Hiromi Ito, Hiroyoshi Nishikawa, Toshio Kitamura, Akinori Kanai, Akihiko Yokoyama, Tohru Fujiwara, Susumu Goyama, Hideki Noguchi, Stanley C. Lee, Atsushi Toyoda, Kunihiko Hinohara, Omar Abdel-Wahab, Daichi Inoue. BRD9 determines the cell fate of hematopoietic stem cells by regulating chromatin state. Nature Communications 14, (2023). doi:10.1038/s41467-023-44081-6
Osamu Miura, Atsushi Toyoda, Tetsuya Sakurai. Chromosome-Scale Genome Assembly of the Freshwater Snail Semisulcospira habei from the Lake Biwa Drainage System. Genome Biology and Evolution 15, (2023). doi:10.1093/gbe/evad208
Bisei Ohkawara, Masaomi Kurokawa, Akinori Kanai, Kiyomi Imamura, Guiying Chen, Ruchen Zhang, Akio Masuda, Koichi Higashi, Hiroshi Mori, Yutaka Suzuki, Ken Kurokawa, Kinji Ohno. Transcriptome profile of subsynaptic myonuclei at the neuromuscular junction in embryogenesis. Journal of Neurochemistry 168, 342-354(2023). doi:10.1111/jnc.16013
Mizue Anda, Shun Yamanouchi, Salvatore Cosentino, Mitsuo Sakamoto, Moriya Ohkuma, Masako Takashima, Atsushi Toyoda, Wataru Iwasaki. Bacteria can maintain rRNA operons solely on plasmids for hundreds of millions of years. Nature Communications 14, (2023). doi:10.1038/s41467-023-42681-w
Masaomi Kurokawa, Koichi Higashi, Keisuke Yoshida, Tomohiko Sato, Shigenori Maruyama, Hiroshi Mori, Ken Kurokawa. Metagenomic Thermometer. DNA Research 30, (2023). doi:10.1093/dnares/dsad024
Masaru Ito, Asako Furukohri, Kenichiro Matsuzaki, Yurika Fujita, Atsushi Toyoda, Akira Shinohara. FIGNL1 AAA+ ATPase remodels RAD51 and DMC1 filaments in pre-meiotic DNA replication and meiotic recombination. Nature Communications 14, (2023). doi:10.1038/s41467-023-42576-w
Kayoko Yamamoto, Ryo Matsuzaki, Wuttipong Mahakham, Wirawan Heman, Hiroyuki Sekimoto, Masanobu Kawachi, Yohei Minakuchi, Atsushi Toyoda, Hisayoshi Nozaki. Expanded male sex-determining region conserved during the evolution of homothallism in the green alga Volvox. iScience 26, 107836(2023). doi:10.1016/j.isci.2023.107836
Kohei Nagao, Yoshiki Tanaka, Rei Kajitani, Atsushi Toyoda, Takehiko Itoh, Souichirou Kubota, Yuji Goto. Bioinformatic and fine-scale chromosomal mapping reveal the nature and evolution of eliminated chromosomes in the Japanese hagfish, Eptatretus burgeri, through analysis of repetitive DNA families. PLOS ONE 18, e0286941(2023). doi:10.1371/journal.pone.0286941
Minoru Moriyama, Yudai Nishide, Atsushi Toyoda, Takehiko Itoh, Takema Fukatsu. Complete genomes of mutualistic bacterial co-symbionts “Candidatus Sulcia muelleri” and “Candidatus Nasuia deltocephalinicola” of the rice green leafhopper Nephotettix cincticeps. Microbiology Resource Announcements 12, (2023). doi:10.1128/MRA.00353-23
Desmila Idola, Hiroshi Mori, Yuji Nagata, Lisa Nonaka, Hirokazu Yano. Host range of strand-biased circularizing integrative elements: a new class of mobile DNA elements nesting in Gammaproteobacteria. Mobile DNA 14, (2023). doi:10.1186/s13100-023-00295-5
Akinobu Toyoda, Yusuke Shibata, Yuzy Matsuo, Kumi Terada, Hiroki Sugimoto, Koichi Higashi, Hiroshi Mori, Akinori Ikeuchi, Masakazu Ito, Ken Kurokawa, Satoshi Katahira. Diversity and compositional differences of the airborne microbiome in a biophilic indoor environment. Scientific Reports 13, (2023). doi:10.1038/s41598-023-34928-9
Taro Onuma, Toshitaka Yamauchi, Hina Kosakamoto, Hibiki Kadoguchi, Takayuki Kuraishi, Takumi Murakami, Hiroshi Mori, Masayuki Miura, Fumiaki Obata. Recognition of commensal bacterial peptidoglycans defines Drosophila gut homeostasis and lifespan. PLOS Genetics 19, e1010709(2023). doi:10.1371/journal.pgen.1010709
Taku Sasaki, Kae Kato, Aoi Hosaka, Yu Fu, Atsushi Toyoda, Asao Fujiyama, Yoshiaki Tarutani, Tetsuji Kakutani. Arms race between anti‐silencing and
Masahiro Oka, Mayumi Otani, Yoichi Miyamoto, Rieko Oshima, Jun Adachi, Takeshi Tomonaga, Munehiro Asally, Yuya Nagaoka, Kaori Tanaka, Atsushi Toyoda, Kazuki Ichikawa, Shinichi Morishita, Kyoichi Isono, Haruhiko Koseki, Ryuichiro Nakato, Yasuyuki Ohkawa, Yoshihiro Yoneda. Phase-separated nuclear bodies of nucleoporin fusions promote condensation of MLL1/CRM1 and rearrangement of 3D genome structure. Cell Reports 42, 112884(2023). doi:10.1016/j.celrep.2023.112884
Takahiro Yamabe, Rei Kajitani, Atsushi Toyoda, Takehiko Itoh. Chromosomal-level genome assembly of the coffee bee hawk moth reveals the evolution of chromosomes and the molecular basis of distinct phenotypes. Genome Biology and Evolution , (2023). doi:10.1093/gbe/evad141
Keiko Shimojima Yamamoto, Takeaki Tamura, Nobuhiko Okamoto, Eriko Nishi, Atsuko Noguchi, Ikuko Takahashi, Yukio Sawaishi, Masaki Shimizu, Hitoshi Kanno, Yohei Minakuchi, Atsushi Toyoda, Toshiyuki Yamamoto. Identification of small-sized intrachromosomal segments at the ends of INV–DUP–DEL patterns. Journal of Human Genetics , (2023). doi:10.1038/s10038-023-01181-x
Vo Phuoc Tuan, Koji Yahara, Ho Dang Quy Dung, Tran Thanh Binh, Pham Huu Tung, Tran Dinh Tri, Ngo Phuong Minh Thuan, Vu Van Khien, Tran Thi Huyen Trang, Bui Hoang Phuc, Evariste Tshibangu-Kabamba, Takashi Matsumoto, Junko Akada, Rumiko Suzuki, Tadayoshi Okimoto, Masaaki Kodama, Kazunari Murakami, Hirokazu Yano, Masaki Fukuyo, Noriko Takahashi, Mototsugu Kato, Shin Nishiumi, Takashi Azuma, Yoshitoshi Ogura, Tetsuya Hayashi, Atsushi Toyoda, Ichizo Kobayashi, Yoshio Yamaoka. Corrigendum: ‘Genome-wide association study of gastric cancer- and duodenal ulcer-derived Helicobacter pylori strains reveals discriminatory genetic variations and novel oncoprotein candidates’. Microbial Genomics 9, (2023). doi:10.1099/mgen.0.001062
Yawako W Kawaguchi, Yuki Tsuchikane, Keisuke Tanaka, Teruaki Taji, Yutaka Suzuki, Atsushi Toyoda, Motomi Ito, Yasuyuki Watano, Tomoaki Nishiyama, Hiroyuki Sekimoto, Takashi Tsuchimatsu. Extensive Copy Number Variation Explains Genome Size Variation in the Unicellular Zygnematophycean Alga, Closterium peracerosum–strigosum–littorale Complex. Genome Biology and Evolution 15, (2023). doi:10.1093/gbe/evad115
Kayoko Yamamoto, Ryo Matsuzaki, Wuttipong Mahakham, Wirawan Heman, Hiroyuki Sekimoto, Masanobu Kawachi, Yohei Minakuchi, Atsushi Toyoda, Hisayoshi Nozaki. Expanded male sex-determining region conserved during the evolution of homothallism in the green alga Volvox. iScience 26, 106893(2023). doi:10.1016/j.isci.2023.106893
Hiroshi Mori, Tamotsu Kato, Hiroaki Ozawa, Mitsuo Sakamoto, Takumi Murakami, Todd D Taylor, Atsushi Toyoda, Moriya Ohkuma, Ken Kurokawa, Hiroshi Ohno. Assessment of metagenomic workflows using a newly constructed human gut microbiome mock community. DNA Research 30, (2023). doi:10.1093/dnares/dsad010
Akira Mabuchi, Shoji Hata, Mariya Genova, Chiharu Tei, Kei K. Ito, Masayasu Hirota, Takuma Komori, Masamitsu Fukuyama, Takumi Chinen, Atsushi Toyoda, Daiju Kitagawa. ssDNA is not superior to dsDNA as long HDR donors for CRISPR-mediated endogenous gene tagging in human diploid RPE1 and HCT116 cells. BMC Genomics 24, (2023). doi:10.1186/s12864-023-09377-3
Kim Nhung Ta, Sae Shimizu‐Sato, Ayumi Agata, Yuri Yoshida, Ken‐ichiro Taoka, Hiroyuki Tsuji, Takashi Akagi, Yasuhiro Tanizawa, Ryosuke Sano, Misuzu Nosaka‐Takahashi, Toshiya Suzuki, Taku Demura, Atsushi Toyoda, Yasukazu Nakamura, Yutaka Sato. A leaf‐emanated signal orchestrates grain size and number in response to maternal resources. The Plant Journal 115, 175-189(2023). doi:10.1111/tpj.16219
Hiroyuki Tanaka, Tatsuki Hori, Shohei Yamamoto, Atsushi Toyoda, Kentaro Yano, Kyoko Yamane, Takehiko Itoh. Haplotype-resolved chromosomal-level assembly of wasabi (Eutrema japonicum) genome. Scientific Data 10, (2023). doi:10.1038/s41597-023-02356-z
Hiroki Ayabe, Atsushi Toyoda, Akitoshi Iwamoto, Nobuhiro Tsutsumi, Shin-ichi Arimura. Mitochondrial gene defects in Arabidopsis can broadly affect mitochondrial gene expression through copy number. Plant Physiology 191, 2256-2275(2023). doi:10.1093/plphys/kiad024
Masaharu Tsuji, Jun-ichi Ishihara, Atsushi Toyoda, Hiroki Takahashi, Sakae Kudoh. Genome Sequence of Basidiomycetous Yeast Mrakia gelida MGH-2, Isolated from Skarvsnes Ice-Free Area, East Antarctica. Microbiology Resource Announcements 12, (2023). doi:10.1128/mra.01064-22
Kim Nhung Ta, Mari W Yoshida, Takumi Tezuka, Sae Shimizu-Sato, Misuzu Nosaka-Takahashi, Atsushi Toyoda, Takamasa Suzuki, Gohta Goshima, Yutaka Sato. Control of Plant Cell Growth and Proliferation by MO25A, a Conserved Major Component of the Mammalian Sterile 20–Like Kinase Pathway. Plant and Cell Physiology 64, 336-351(2023). doi:10.1093/pcp/pcad005
Eri Sakamoto, Yasuhiro Katahira, Izuru Mizoguchi, Aruma Watanabe, Yuma Furusaka, Ami Sekine, Miu Yamagishi, Jukito Sonoda, Satomi Miyakawa, Shinya Inoue, Hideaki Hasegawa, Kazuyuki Yo, Fumiya Yamaji, Akemi Toyoda, Takayuki Yoshimoto. Chemical- and Drug-Induced Allergic, Inflammatory, and Autoimmune Diseases Via Haptenation. Biology 12, 123(2023). doi:10.3390/biology12010123
Hiroyuki Sekimoto, Ayumi Komiya, Natsumi Tsuyuki, Junko Kawai, Naho Kanda, Ryo Ootsuki, Yutaka Suzuki, Atsushi Toyoda, Asao Fujiyama, Masahiro Kasahara, Jun Abe, Yuki Tsuchikane, Tomoaki Nishiyama. A divergent
Kenta Shirasawa, Munetaka Hosokawa, Yasuo Yasui, Atsushi Toyoda, Sachiko Isobe. Chromosome-scale genome assembly of a Japanese chili pepper landrace, Capsicum annuum ‘Takanotsume’. DNA Research 30, (2022). doi:10.1093/dnares/dsac052
Takeaki Tamura, Keiko Yamamoto Shimojima, Nobuhiko Okamoto, Hiroshi Yagasaki, Ichiro Morioka, Hitoshi Kanno, Yohei Minakuchi, Atsushi Toyoda, Toshiyuki Yamamoto. Long‐read sequence analysis for clustered genomic copy number aberrations revealed architectures of intricately intertwined rearrangements. American Journal of Medical Genetics Part A 191, 112-119(2022). doi:10.1002/ajmg.a.62997
Masaru Konishi Nobu, Ryosuke Nakai, Satoshi Tamazawa, Hiroshi Mori, Atsushi Toyoda, Akira Ijiri, Shino Suzuki, Ken Kurokawa, Yoichi Kamagata, Hideyuki Tamaki. Unique H2-utilizing lithotrophy in serpentinite-hosted systems. The ISME Journal 17, 95-104(2022). doi:10.1038/s41396-022-01197-9
Tetsuo Kon, Kentaro Fukuta, Zelin Chen, Koto Kon-Nanjo, Kota Suzuki, Masakazu Ishikawa, Hikari Tanaka, Shawn M. Burgess, Hideki Noguchi, Atsushi Toyoda, Yoshihiro Omori. Single-cell transcriptomics of the goldfish retina reveals genetic divergence in the asymmetrically evolved subgenomes after allotetraploidization. Communications Biology 5, (2022). doi:10.1038/s42003-022-04351-3
Amit Rai, Hideki Hirakawa, Megha Rai, Yohei Shimizu, Kenta Shirasawa, Shinji Kikuchi, Hikaru Seki, Mami Yamazaki, Atsushi Toyoda, Sachiko Isobe, Toshiya Muranaka, Kazuki Saito. Chromosome-scale genome assembly of Glycyrrhiza uralensis revealed metabolic gene cluster centred specialized metabolites biosynthesis. DNA Research 29, (2022). doi:10.1093/dnares/dsac043
Hiroyuki Kato, Keisuke Tateishi, Dosuke Iwadate, Keisuke Yamamoto, Hiroaki Fujiwara, Takuma Nakatsuka, Yotaro Kudo, Yoku Hayakawa, Hideaki Ijichi, Motoyuki Otsuka, Takahiro Kishikawa, Ryota Takahashi, Koji Miyabayashi, Yousuke Nakai, Yoshihiro Hirata, Atsushi Toyoda, Shinichi Morishita, Mitsuhiro Fujishiro. HNF1B‐driven three‐dimensional chromatin structure for molecular classification in pancreatic cancers. Cancer Science 114, 1672-1685(2022). doi:10.1111/cas.15690
Miho Terao, Yuya Ogawa, Shuji Takada, Rei Kajitani, Miki Okuno, Yuta Mochimaru, Kentaro Matsuoka, Takehiko Itoh, Atsushi Toyoda, Tomohiro Kono, Takamichi Jogahara, Shusei Mizushima, Asato Kuroiwa. Turnover of mammal sex chromosomes in the Sry-deficient Amami spiny rat is due to male-specific upregulation ofSox9. Proceedings of the National Academy of Sciences 119, (2022). doi:10.1073/pnas.2211574119
Kana Morinaga, Hiroyuki Kusada, Sachiko Sakamoto, Takumi Murakami, Atsushi Toyoda, Hiroshi Mori, Xian-Ying Meng, Motoko Takashino, Kazutoshi Murotomi, Hideyuki Tamaki. Granulimonas faecalis gen. nov., sp. nov., and Leptogranulimonas caecicola gen. nov., sp. nov., novel lactate-producing Atopobiaceae bacteria isolated from mouse intestines, and an emended description of the family Atopobiaceae. International Journal of Systematic and Evolutionary Microbiology 72, (2022). doi:10.1099/ijsem.0.005596
Yukari Tanaka, Riu Yamashita, Junko Kawashima, Hiroshi Mori, Ken Kurokawa, Shinji Fukuda, Yasuhiro Gotoh, Keiji Nakamura, Tetsuya Hayashi, Yoshiyuki Kasahara, Yukuto Sato, Shin Fukudo. Omics profiles of fecal and oral microbiota change in irritable bowel syndrome patients with diarrhea and symptom exacerbation. Journal of Gastroenterology 57, 748-760(2022). doi:10.1007/s00535-022-01888-2
Masa-aki Yoshida, Kazuki Hirota, Junichi Imoto, Miki Okuno, Hiroyuki Tanaka, Rei Kajitani, Atsushi Toyoda, Takehiko Itoh, Kazuho Ikeo, Takenori Sasaki, Davin H E Setiamarga. Gene Recruitments and Dismissals in the Argonaut Genome Provide Insights into Pelagic Lifestyle Adaptation and Shell-like Eggcase Reacquisition. Genome Biology and Evolution 14, (2022). doi:10.1093/gbe/evac140
Osamu Nishimura, John Rozewicki, Kazuaki Yamaguchi, Kaori Tatsumi, Yuta Ohishi, Tazro Ohta, Masaru Yagura, Taiki Niwa, Chiharu Tanegashima, Akinori Teramura, Shotaro Hirase, Akane Kawaguchi, Milton Tan, Salvatore D'Aniello, Filipe Castro, André Machado, Mitsumasa Koyanagi, Akihisa Terakita, Ryo Misawa, Masayuki Horie, Junna Kawasaki, Takashi Asahida, Atsuko Yamaguchi, Kiyomi Murakumo, Rui Matsumoto, Iker Irisarri, Norio Miyamoto, Atsushi Toyoda, Sho Tanaka, Tatsuya Sakamoto, Yasuko Semba, Shinya Yamauchi, Kazuyuki Yamada, Kiyonori Nishida, Itsuki Kiyatake, Keiichi Sato, Susumu Hyodo, Mitsutaka Kadota, Yoshinobu Uno, Shigehiro Kuraku. Squalomix: shark and ray genome analysis consortium and its data sharing platform. F1000Research 11, 1077(2022). doi:10.12688/f1000research.123591.1
Jessica Kim, Masafumi Muraoka, Hajime Okada, Atsushi Toyoda, Rieko Ajima, Yumiko Saga. The RNA helicase DDX6 controls early mouse embryogenesis by repressing aberrant inhibition of BMP signaling through miRNA-mediated gene silencing. PLOS Genetics 18, e1009967(2022). doi:10.1371/journal.pgen.1009967
Masaharu Tsuji, Jun-ichi Ishihara, Atsushi Toyoda, Hiroki Takahashi. High-Quality Genome Sequence of Cystobasidium tubakii JCM 31526 T, Isolated from East Ongul Island, Antarctica. Microbiology Resource Announcements 11, (2022). doi:10.1128/mra.00741-22
Kyosuke Yakabe, Seiichiro Higashi, Masahiro Akiyama, Hiroshi Mori, Takumi Murakami, Atsushi Toyoda, Yuta Sugiyama, Shigenobu Kishino, Kenji Okano, Akiyoshi Hirayama, Aina Gotoh, Shunyi Li, Takeshi Mori, Takane Katayama, Jun Ogawa, Shinji Fukuda, Koji Hase, Yun-Gi Kim. Dietary-protein sources modulate host susceptibility to Clostridioides difficile infection through the gut microbiota. Cell Reports 40, 111332(2022). doi:10.1016/j.celrep.2022.111332
Mika Uehara, Takashi Inoue, Minori Kominato, Sumitaka Hase, Erika Sasaki, Atsushi Toyoda, Yasubumi Sakakibara. Intraintestinal Analysis of the Functional Activity of Microbiomes and Its Application to the Common Marmoset Intestine. mSystems 7, (2022). doi:10.1128/msystems.00520-22
Daisuke Sekine, Satoshi Oku, Tsukasa Nunome, Hideki Hirakawa, Mai Tsujimura, Toru Terachi, Atsushi Toyoda, Masayoshi Shigyo, Shusei Sato, Hikaru Tsukazaki. Development of a genome-wide marker design workflow for onions and its application in target amplicon sequencing-based genotyping. DNA Research 29, (2022). doi:10.1093/dnares/dsac020
Yuma Koizumi, Junzo Hisatsune, Atsushi Toyoda, Osamu Yamasaki, Jun Kaneko, Motoyuki Sugai. Complete Genome Sequence of a Hyaluronate Lyase HysA- and HysB-Producing, Methicillin-Resistant Staphylococcus aureus Sequence Type 30, Staphylococcal Cassette ChromosomemecType IVc Strain Isolated from Furunculosis in Japan. Microbiology Resource Announcements 11, (2022). doi:10.1128/mra.00308-22
Yusuke Okazaki, Shin-ichi Nakano, Atsushi Toyoda, Hideyuki Tamaki. Long-Read-Resolved, Ecosystem-Wide Exploration of Nucleotide and Structural Microdiversity of Lake Bacterioplankton Genomes. mSystems 7, (2022). doi:10.1128/msystems.00433-22
Takahiko Kubo, Yoshiyuki Yamagata, Hiroaki Matsusaka, Atsushi Toyoda, Yutaka Sato, Toshihiro Kumamaru. Whole-Genome Sequencing of Rice Mutant Library Members Induced by N-Methyl-N-Nitrosourea Mutagenesis of Fertilized Egg Cells. Rice 15, (2022). doi:10.1186/s12284-022-00585-1
Yasuhiko Chikami, Miki Okuno, Atsushi Toyoda, Takehiko Itoh, Teruyuki Niimi. Evolutionary History of Sexual Differentiation Mechanism in Insects. Molecular Biology and Evolution 39, (2022). doi:10.1093/molbev/msac145
Takumi Kamiyama, Yuko Shimada-Niwa, Hiroyuki Tanaka, Minami Katayama, Takayoshi Kuwabara, Hitoha Mori, Akari Kunihisa, Takehiko Itoh, Atsushi Toyoda, Ryusuke Niwa. Whole-genome sequencing analysis and protocol for RNA interference of the endoparasitoid wasp Asobara japonica. DNA Research 29, (2022). doi:10.1093/dnares/dsac019
Ahammad Kabir, Risa Ieda, Sho Hosoya, Daigaku Fujikawa, Kazufumi Atsumi, Shota Tajima, Aoi Nozawa, Takashi Koyama, Shotaro Hirase, Osamu Nakamura, Mitsutaka Kadota, Osamu Nishimura, Shigehiro Kuraku, Yasukazu Nakamura, Hisato Kobayashi, Atsushi Toyoda, Satoshi Tasumi, Kiyoshi Kikuchi. Repeated translocation of a supergene underlying rapid sex chromosome turnover in Takifugu pufferfish. Proceedings of the National Academy of Sciences 119, (2022). doi:10.1073/pnas.2121469119
Hidefumi Suzuki, Ryota Abe, Miho Shimada, Tomonori Hirose, Hiroko Hirose, Keisuke Noguchi, Yoko Ike, Nanami Yasui, Kazuki Furugori, Yuki Yamaguchi, Atsushi Toyoda, Yutaka Suzuki, Tatsuro Yamamoto, Noriko Saitoh, Shigeo Sato, Chieri Tomomori-Sato, Ronald C. Conaway, Joan W. Conaway, Hidehisa Takahashi. The 3′ Pol II pausing at replication-dependent histone genes is regulated by Mediator through Cajal bodies’ association with histone locus bodies. Nature Communications 13, (2022). doi:10.1038/s41467-022-30632-w
Qiushi Li, Dorothea Lindtke, Carlos Rodríguez-Ramírez, Ryo Kakioka, Hiroshi Takahashi, Atsushi Toyoda, Jun Kitano, Rachel L. Ehrlich, Joshua Chang Mell, Sam Yeaman. Local Adaptation and the Evolution of Genome Architecture in Threespine Stickleback. Genome Biology and Evolution 14, (2022). doi:10.1093/gbe/evac075
Arikuni Uchimura, Hirotaka Matsumoto, Yasunari Satoh, Yohei Minakuchi, Sayaka Wakayama, Teruhiko Wakayama, Mayumi Higuchi, Masakazu Hashimoto, Ryutaro Fukumura, Atsushi Toyoda, Yoichi Gondo, Takeshi Yagi. Early embryonic mutations reveal dynamics of somatic and germ cell lineages in mice. Genome Research , (2022). doi:10.1101/gr.276363.121
Yoshimi Yukawa‐Muto, Tomonori Kamiya, Hideki Fujii, Hiroshi Mori, Atsushi Toyoda, Ikuya Sato, Yusuke Konishi, Akiyoshi Hirayama, Eiji Hara, Shinji Fukuda, Norifumi Kawada, Naoko Ohtani. Distinct responsiveness to rifaximin in patients with hepatic encephalopathy depends on functional gut microbial species. Hepatology Communications 6, 2090-2104(2022). doi:10.1002/hep4.1954
Michael T. Madigan, Jill N. Absher, Joseph E. Mayers, Marie Asao, Deborah O. Jung, Kelly S. Bender, Megan L. Kempher, Mackenzie K. Hayward, Sophia A. Sanguedolce, Abigail C. Brown, Shinichi Takaichi, Ken Kurokawa, Atsushi Toyoda, Hiroshi Mori, Yusuke Tsukatani, Zheng-Yu Wang-Otomo, David M. Ward, W. Matthew Sattley. Correction to: Allochromatium tepidum, sp. nov., a hot spring species of purple sulfur bacteria. Archives of Microbiology 204, (2022). doi:10.1007/s00203-022-02827-8
Satoshi Hiraoka, Tomomi Sumida, Miho Hirai, Atsushi Toyoda, Shinsuke Kawagucci, Taichi Yokokawa, Takuro Nunoura. Diverse DNA modification in marine prokaryotic and viral communities. Nucleic Acids Research 50, 1531-1550(2022). doi:10.1093/nar/gkab1292
Michael T. Madigan, Jill N. Absher, Joseph E. Mayers, Marie Asao, Deborah O. Jung, Kelly S. Bender, Megan L. Kempher, Mackenzie K. Hayward, Sophia A. Sanguedolce, Abigail C. Brown, Shinichi Takaichi, Ken Kurokawa, Atsushi Toyoda, Hiroshi Mori, Yusuke Tsukatani, Zheng-Yu Wang-Otomo, David M. Ward, W. Matthew Sattley. Allochromatium tepidum, sp. nov., a hot spring species of purple sulfur bacteria. Archives of Microbiology 204, (2022). doi:10.1007/s00203-021-02715-7
Hiroyuki Kato, Keisuke Tateishi, Hiroaki Fujiwara, Takuma Nakatsuka, Keisuke Yamamoto, Yotaro Kudo, Yoku Hayakawa, Hayato Nakagawa, Yasuo Tanaka, Hideaki Ijichi, Motoyuki Otsuka, Dosuke Iwadate, Hiroki Oyama, Sachiko Kanai, Kensaku Noguchi, Tatsunori Suzuki, Tatsuya Sato, Ryunosuke Hakuta, Kazunaga Ishigaki, Kei Saito, Tomotaka Saito, Naminatsu Takahara, Takahiro Kishikawa, Tsuyoshi Hamada, Ryota Takahashi, Koji Miyabayashi, Suguru Mizuno, Hirofumi Kogure, Yousuke Nakai, Yoshihiro Hirata, Atsushi Toyoda, Kazuki Ichikawa, Wei Qu, Shinichi Morishita, Junichi Arita, Mariko Tanaka, Tetsuo Ushiku, Kiyoshi Hasegawa, Mitsuhiro Fujishiro, Kazuhiko Koike. MNX1-HNF1B Axis Is Indispensable for Intraductal Papillary Mucinous Neoplasm Lineages. Gastroenterology 162, 1272-1287.e16(2022). doi:10.1053/j.gastro.2021.12.254
Shuji Shigenobu, Yoshinobu Hayashi, Dai Watanabe, Gaku Tokuda, Masaru Y. Hojo, Kouhei Toga, Ryota Saiki, Hajime Yaguchi, Yudai Masuoka, Ryutaro Suzuki, Shogo Suzuki, Moe Kimura, Masatoshi Matsunami, Yasuhiro Sugime, Kohei Oguchi, Teruyuki Niimi, Hiroki Gotoh, Masaru K. Hojo, Satoshi Miyazaki, Atsushi Toyoda, Toru Miura, Kiyoto Maekawa. Genomic and transcriptomic analyses of the subterranean termite Reticulitermes speratus : Gene duplication facilitates social evolution. Proceedings of the National Academy of Sciences 119, (2022). doi:10.1073/pnas.2110361119
- 2021年 -
Toyoyuki Takada, Kentaro Fukuta, Daiki Usuda, Tatsuya Kushida, Shinji Kondo, Shoko Kawamoto, Atsushi Yoshiki, Yuichi Obata, Asao Fujiyama, Atsushi Toyoda, Hideki Noguchi, Toshihiko Shiroishi, Hiroshi Masuya. MoG+: a database of genomic variations across three mouse subspecies for biomedical research. Mammalian Genome 33, 31-43(2021). doi:10.1007/s00335-021-09933-w
Tsubasa Kawai, Kyosuke Shibata, Ryosuke Akahoshi, Shunsaku Nishiuchi, Hirokazu Takahashi, Mikio Nakazono, Takaaki Kojima, Misuzu Nosaka-Takahashi, Yutaka Sato, Atsushi Toyoda, Nonawin Lucob-Agustin, Mana Kano-Nakata, Roel R. Suralta, Jonathan M. Niones, Yinglong Chen, Kadambot H. M. Siddique, Akira Yamauchi, Yoshiaki Inukai. WUSCHEL-related homeoboxfamily genes in rice control lateral root primordium size. Proceedings of the National Academy of Sciences 119, (2021). doi:10.1073/pnas.2101846119
Ruriko Nishida, Keiji Nakamura, Itsuki Taniguchi, Kazunori Murase, Tadasuke Ooka, Yoshitoshi Ogura, Yasuhiro Gotoh, Takehiko Itoh, Atsushi Toyoda, Jacques Georges Mainil, Denis Piérard, Kazuko Seto, Tetsuya Harada, Junko Isobe, Keiko Kimata, Yoshiki Etoh, Mitsuhiro Hamasaki, Hiroshi Narimatsu, Jun Yatsuyanagi, Mitsuhiro Kameyama, Yuko Matsumoto, Yuhki Nagai, Jun Kawase, Eiji Yokoyama, Kazuhiko Ishikawa, Takayuki Shiomoto, Kenichi Lee, Dongchon Kang, Koichi Akashi, Makoto Ohnishi, Sunao Iyoda, Tetsuya Hayashi. The global population structure and evolutionary history of the acquisition of major virulence factor-encoding genetic elements in Shiga toxin-producing Escherichia coli O121:H19. Microbial Genomics 7, (2021). doi:10.1099/mgen.0.000716
Mitsuo Sakamoto, Nao Ikeyama, Masahiro Yuki, Takumi Murakami, Hiroshi Mori, Takao Iino, Moriya Ohkuma. Adlercreutzia hattorii sp. nov., an equol non-producing bacterium isolated from human faeces. International Journal of Systematic and Evolutionary Microbiology 71, (2021). doi:10.1099/ijsem.0.005121
Vo Phuoc Tuan, Koji Yahara, Ho Dang Quy Dung, Tran Thanh Binh, Pham Huu Tung, Tran Dinh Tri, Ngo Phuong Minh Thuan, Vu Van Khien, Tran Thi Huyen Trang, Bui Hoang Phuc, Evariste Tshibangu-Kabamba, Takashi Matsumoto, Junko Akada, Rumiko Suzuki, Tadayoshi Okimoto, Masaaki Kodama, Kazunari Murakami, Hirokazu Yano, Masaki Fukuyo, Noriko Takahashi, Mototsugu Kato, Shin Nishiumi, Takashi Azuma, Yoshitoshi Ogura, Tetsuya Hayashi, Atsushi Toyoda, Ichizo Kobayashi, Yoshio Yamaoka. Genome-wide association study of gastric cancer- and duodenal ulcer-derived Helicobacter pylori strains reveals discriminatory genetic variations and novel oncoprotein candidates. Microbial Genomics 7, (2021). doi:10.1099/mgen.0.000680
Khoa Lai, Ngoc Thai Nguyen, Michiko Yasuda, Khondoker M.G. Dastogeer, Atsushi Toyoda, Koichi Higashi, Ken Kurokawa, Nga Thi Thu Nguyen, Ken Komatsu, Shin Okazaki. Leaf Bleaching in Rice: A New Disease in Vietnam Caused by Methylobacterium indicum, Its Genomic Characterization and the Development of a Suitable Detection Technique. Microbes and Environments 36, (2021). doi:10.1264/jsme2.ME21035
Masafumi Nozawa, Yohei Minakuchi, Kazuhiro Satomura, Shu Kondo, Atsushi Toyoda, Koichiro Tamura. Shared evolutionary trajectories of three independent neo-sex chromosomes in Drosophila. Genome Research 31, 2069-2079(2021). doi:10.1101/gr.275503.121
Rei Kajitani, Hideki Noguchi, Yasuhiro Gotoh, Yoshitoshi Ogura, Dai Yoshimura, Miki Okuno, Atsushi Toyoda, Tomomi Kuwahara, Tetsuya Hayashi, Takehiko Itoh. MetaPlatanus: a metagenome assembler that combines long-range sequence links and species-specific features. Nucleic Acids Research 49, e130-e130(2021). doi:10.1093/nar/gkab831
Ixchel F. Mandagi, Ryo Kakioka, Javier Montenegro, Hirozumi Kobayashi, Kawilarang W. A. Masengi, Nobuyuki Inomata, Atsushi J. Nagano, Atsushi Toyoda, Satoshi Ansai, Masatoshi Matsunami, Ryosuke Kimura, Jun Kitano, Junko Kusumi, Kazunori Yamahira. Species divergence and repeated ancient hybridization in a Sulawesian lake system. Journal of Evolutionary Biology 34, 1767-1780(2021). doi:10.1111/jeb.13932
Takehiro Sato, Noboru Adachi, Ryosuke Kimura, Kazuyoshi Hosomichi, Minoru Yoneda, Hiroki Oota, Atsushi Tajima, Atsushi Toyoda, Hideaki Kanzawa-Kiriyama, Hiromi Matsumae, Kae Koganebuchi, Kentaro K Shimizu, Ken-Ichi Shinoda, Tsunehiko Hanihara, Andrzej Weber, Hirofumi Kato, Hajime Ishida. Whole Genome Sequencing of a 900-year-old Human Skeleton Supports Two Past Migration Events from the Russian Far East to Northern Japan. Genome Biology and Evolution , (2021). doi:10.1093/gbe/evab192
Hideaki Yuasa, Rei Kajitani, Yuta Nakamura, Kazuki Takahashi, Miki Okuno, Fumiya Kobayashi, Takahiro Shinoda, Atsushi Toyoda, Yutaka Suzuki, Nalinee Thongtham, Zac Forsman, Omri Bronstein, Davide Seveso, Enrico Montalbetti, Coralie Taquet, Gal Eyal, Nina Yasuda, Takehiko Itoh. Elucidation of the speciation history of three sister species of crown-of-thorns starfish (Acanthaster spp.) based on genomic analysis. DNA Research 28, (2021). doi:10.1093/dnares/dsab012
Nozomi Yamamoto, Naoki Watarai, Hitoshi Koyano, Kazunori Sawada, Atsushi Toyoda, Ken Kurokawa, Takuji Yamada. Analysis of genomic characteristics and their influence on metabolism in Aspergillus luchuensis albino mutants using genome sequencing. Fungal Genetics and Biology 155, 103601(2021). doi:10.1016/j.fgb.2021.103601
Hideki Hirakawa, Atsushi Toyoda, Takehiko Itoh, Yutaka Suzuki, Atsushi J Nagano, Suguru Sugiyama, Yasuyuki Onodera. A spinach genome assembly with remarkable completeness, and its use for rapid identification of candidate genes for agronomic traits. DNA Research 28, (2021). doi:10.1093/dnares/dsab004
Guillem Ylla, Taro Nakamura, Takehiko Itoh, Rei Kajitani, Atsushi Toyoda, Sayuri Tomonari, Tetsuya Bando, Yoshiyasu Ishimaru, Takahito Watanabe, Masao Fuketa, Yuji Matsuoka, Austen A. Barnett, Sumihare Noji, Taro Mito, Cassandra G. Extavour. Insights into the genomic evolution of insects from cricket genomes. Communications Biology 4, (2021). doi:10.1038/s42003-021-02197-9
Keiichiro Koiwai, Takashi Koyama, Soichiro Tsuda, Atsushi Toyoda, Kiyoshi Kikuchi, Hiroaki Suzuki, Ryuji Kawano. Single-cell RNA-seq analysis reveals penaeid shrimp hemocyte subpopulations and cell differentiation process. eLife 10, (2021). doi:10.7554/eLife.66954
Taisei Kikuchi, Mehmet Dayi, Vicky L. Hunt, Kenji Ishiwata, Atsushi Toyoda, Asuka Kounosu, Simo Sun, Yasunobu Maeda, Yoko Kondo, Belkisyole Alarcon de Noya, Oscar Noya, Somei Kojima, Toshiaki Kuramochi, Haruhiko Maruyama. Genome of the fatal tapeworm Sparganum proliferum uncovers mechanisms for cryptic life cycle and aberrant larval proliferation. Communications Biology 4, (2021). doi:10.1038/s42003-021-02160-8
Ranna Nakao, Kentaro Kasama, Bazartseren Boldbaatar, Yoshitoshi Ogura, Hiroki Kawabata, Atsushi Toyoda, Tetsuya Hayashi, Ai Takano, Ken Maeda. The evolution of hard tick-borne relapsing fever borreliae is correlated with vector species rather than geographical distance. BMC Ecology and Evolution 21, (2021). doi:10.1186/s12862-021-01838-1
Kayoko Yamamoto, Takashi Hamaji, Hiroko Kawai-Toyooka, Ryo Matsuzaki, Fumio Takahashi, Yoshiki Nishimura, Masanobu Kawachi, Hideki Noguchi, Yohei Minakuchi, James G. Umen, Atsushi Toyoda, Hisayoshi Nozaki. Three genomes in the algal genus Volvox reveal the fate of a haploid sex-determining region after a transition to homothallism. Proceedings of the National Academy of Sciences 118, e2100712118(2021). doi:10.1073/pnas.2100712118
Mitsuo Sakamoto, Nao Ikeyama, Atsushi Toyoda, Takumi Murakami, Hiroshi Mori, Sho Morohoshi, Tadao Kunihiro, Takao Iino, Moriya Ohkuma. Coprobacter secundus subsp.similis subsp. nov. and Solibaculum mannosilyticum gen. nov., sp. nov., isolated from human feces. Microbiology and Immunology 65, 245-256(2021). doi:10.1111/1348-0421.12886
Kazuki Hirota, Masa-aki Yoshida, Takehiko Itoh, Atsushi Toyoda, Davin H. E. Setiamarga. The full mitochondrial genome sequence of the greater argonaut Argonauta argo (Cephalopoda, Argonautoidea) and its phylogenetic position in Octopodiformes. Mitochondrial DNA Part B 6, 1451-1453(2021). doi:10.1080/23802359.2021.1911710
Taro Maeda, Shunichi Takahashi, Takao Yoshida, Shigeru Shimamura, Yoshihiro Takaki, Yukiko Nagai, Atsushi Toyoda, Yutaka Suzuki, Asuka Arimoto, Hisaki Ishii, Nori Satoh, Tomoaki Nishiyama, Mitsuyasu Hasebe, Tadashi Maruyama, Jun Minagawa, Junichi Obokata, Shuji Shigenobu. Chloroplast acquisition without the gene transfer in kleptoplastic sea slugs, Plakobranchus ocellatus. eLife 10, (2021). doi:10.7554/eLife.60176
Yasuyuki Yamada, Hideki Hirakawa, Kentaro Hori, Yohei Minakuchi, Atsushi Toyoda, Nobukazu Shitan, Fumihiko Sato. Comparative analysis using the draft genome sequence of California poppy (Eschscholzia californica) for exploring the candidate genes involved in benzylisoquinoline alkaloid biosynthesis. Bioscience, Biotechnology, and Biochemistry 85, 851-859(2021). doi:10.1093/bbb/zbaa091
Misako Yajima, Risako Kakuta, Yutaro Saito, Shiori Kitaya, Atsushi Toyoda, Kazufumi Ikuta, Jun Yasuda, Nobuo Ohta, Teru Kanda. A global phylogenetic analysis of Japanese tonsil-derived Epstein–Barr virus strains using viral whole-genome cloning and long-read sequencing. Journal of General Virology 102, (2021). doi:10.1099/jgv.0.001549
Hitomi Takei, Kenta Shirasawa, Kosuke Kuwabara, Atsushi Toyoda, Yuma Matsuzawa, Shinji Iioka, Tohru Ariizumi. De novo genome assembly of two tomato ancestors, Solanum pimpinellifolium and Solanum lycopersicum var. cerasiforme, by long-read sequencing. DNA Research 28, (2021). doi:10.1093/dnares/dsaa029
Amit Rai, Hideki Hirakawa, Ryo Nakabayashi, Shinji Kikuchi, Koki Hayashi, Megha Rai, Hiroshi Tsugawa, Taiki Nakaya, Tetsuya Mori, Hideki Nagasaki, Runa Fukushi, Yoko Kusuya, Hiroki Takahashi, Hiroshi Uchiyama, Atsushi Toyoda, Shoko Hikosaka, Eiji Goto, Kazuki Saito, Mami Yamazaki. Chromosome-level genome assembly of Ophiorrhiza pumila reveals the evolution of camptothecin biosynthesis. Nature Communications 12, (2021). doi:10.1038/s41467-020-20508-2
Haruna Nakamura, Mitsuto Aibara, Rei Kajitani, Hillary D J Mrosso, Semvua I Mzighani, Atsushi Toyoda, Takehiko Itoh, Norihiro Okada, Masato Nikaido. Genomic Signatures for Species-Specific Adaptation in Lake Victoria Cichlids Derived from Large-Scale Standing Genetic Variation. Molecular Biology and Evolution , (2021). doi:10.1093/molbev/msab084
Yuki Matsumoto, Hiromichi Nagayama, Hirofumi Nakaoka, Atsushi Toyoda, Tatsuhiko Goto, Tsuyoshi Koide. Combined change of behavioral traits for domestication and gene‐networks in mice selectively bred for active tameness. Genes, Brain and Behavior 20, (2021). doi:10.1111/gbb.12721
Yukako Katsura, Toshimichi Ikemura, Rei Kajitani, Atsushi Toyoda, Takehiko Itoh, Mitsuaki Ogata, Ikuo Miura, Kennosuke Wada, Yoshiko Wada, Yoko Satta. Comparative genomics of Glandirana rugosa using unsupervised AI reveals a high CG frequency. Life Science Alliance 4, e202000905(2021). doi:10.26508/lsa.202000905
Hiromi Kajiya-Kanegae, Hajime Ohyanagi, Toshinobu Ebata, Yasuhiro Tanizawa, Akio Onogi, Yuji Sawada, Masami Yokota Hirai, Zi-Xuan Wang, Bin Han, Atsushi Toyoda, Asao Fujiyama, Hiroyoshi Iwata, Katsutoshi Tsuda, Toshiya Suzuki, Misuzu Nosaka-Takahashi, Ken-ichi Nonomura, Yasukazu Nakamura, Shoko Kawamoto, Nori Kurata, Yutaka Sato. OryzaGenome2.1: Database of Diverse Genotypes in Wild Oryza Species. Rice 14, (2021). doi:10.1186/s12284-021-00468-x
Kyoko Ishino, Hidetoshi Hasuwa, Jun Yoshimura, Yuka W Iwasaki, Hidenori Nishihara, Naomi M Seki, Takamasa Hirano, Marie Tsuchiya, Hinako Ishizaki, Harumi Masuda, Tae Kuramoto, Kuniaki Saito, Yasubumi Sakakibara, Atsushi Toyoda, Takehiko Itoh, Mikiko C Siomi, Shinichi Morishita, Haruhiko Siomi. Hamster PIWI proteins bind to piRNAs with stage-specific size variations during oocyte maturation. Nucleic Acids Research 49, 2700-2720(2021). doi:10.1093/nar/gkab059
Koji Inoue, Yuki Yoshioka, Hiroyuki Tanaka, Azusa Kinjo, Mieko Sassa, Ikuo Ueda, Chuya Shinzato, Atsushi Toyoda, Takehiko Itoh. Genomics and transcriptomics of the green mussel explain the durability of its byssus.Scientific Reports 11, (2021). doi:10.1038/s41598-021-84948-6
Satoshi Ansai, Koji Mochida, Shingo Fujimoto, Daniel F. Mokodongan, Bayu Kreshna Adhitya Sumarto, Kawilarang W. A. Masengi, Renny K. Hadiaty, Atsushi J. Nagano, Atsushi Toyoda, Kiyoshi Naruse, Kazunori Yamahira, Jun Kitano. Genome editing reveals fitness effects of a gene for sexual dichromatism in Sulawesian fishes. Nature Communications 12, (2021). doi:10.1038/s41467-021-21697-0
- 2020年 -
Takashi Okubo, Atsushi Toyoda, Kohei Fukuhara, Ikuo Uchiyama, Yuhki Harigaya, Megumi Kuroiwa, Takuma Suzuki, Yuka Murakami, Yuichi Suwa, Hideto Takami. The physiological potential of anammox bacteria as revealed by their core genome structure. DNA Research 28, (2020). doi:10.1093/dnares/dsaa028
Taiko Kim To, Yuichiro Nishizawa, Soichi Inagaki, Yoshiaki Tarutani, Sayaka Tominaga, Atsushi Toyoda, Asao Fujiyama, Frédéric Berger, Tetsuji Kakutani. Author Correction: RNA interference-independent reprogramming of DNA methylation in Arabidopsis. Nature Plants 7, 233-233(2020). doi:10.1038/s41477-020-00839-0
Mitsuo Sakamoto, Nao Ikeyama, Atsushi Toyoda, Takumi Murakami, Hiroshi Mori, Moriya Ohkuma. Complete Genome Sequence of Faecalibacillus intestinalis JCM 34082, Isolated from Feces from a Healthy Japanese Female. Microbiology Resource Announcements 9, (2020). doi:10.1128/MRA.01160-20
Taiko Kim To, Yuichiro Nishizawa, Soichi Inagaki, Yoshiaki Tarutani, Sayaka Tominaga, Atsushi Toyoda, Asao Fujiyama, Frédéric Berger, Tetsuji Kakutani. RNA interference-independent reprogramming of DNA methylation in Arabidopsis. Nature Plants 6, 1455-1467(2020). doi:10.1038/s41477-020-00810-z
Ryo Kakioka, Seiichi Mori, Tomoyuki Kokita, Takuya K. Hosoki, Atsushi J. Nagano, Asano Ishikawa, Manabu Kume, Atsushi Toyoda, Jun Kitano. Multiple waves of freshwater colonization of the three-spined stickleback in the Japanese Archipelago. BMC Evolutionary Biology 20, (2020). doi:10.1186/s12862-020-01713-5
Songkui Cui, Tomoya Kubota, Tomoaki Nishiyama, Juliane K. Ishida, Shuji Shigenobu, Tomoko F. Shibata, Atsushi Toyoda, Mitsuyasu Hasebe, Ken Shirasu, Satoko Yoshida. Ethylene signaling mediates host invasion by parasitic plants. Science Advances 6, eabc2385(2020). doi:10.1126/sciadv.abc2385
Shin‐ichi Arimura, Hiroki Ayabe, Hajime Sugaya, Miki Okuno, Yoshiko Tamura, Yu Tsuruta, Yuta Watari, Shungo Yanase, Takaki Yamauchi, Takehiko Itoh, Atsushi Toyoda, Hideki Takanashi, Nobuhiro Tsutsumi. Targeted gene disruption of ATP synthases 6‐1 and 6‐2 in the mitochondrial genome of Arabidopsis thaliana by mitoTALENs. The Plant Journal 104, 1459-1471(2020). doi:10.1111/tpj.15041
Yoshiteru Noutoshi, Atsushi Toyoda, Tomoya Ishii, Kirara Saito, Megumi Watanabe, Akira Kawaguchi. Complete Genome Sequence Data of Nonpathogenic Strain Rhizobium vitis VAR03-1, a Biological Control Agent for Grapevine Crown Gall Disease. Molecular Plant-Microbe Interactions® 33, 1451-1453(2020). doi:10.1094/MPMI-07-20-0181-A
Yoshiteru Noutoshi, Atsushi Toyoda, Tomoya Ishii, Kirara Saito, Megumi Watanabe, Akira Kawaguchi. Complete Genome Sequence Data of Tumorigenic Rhizobium vitis Strain VAT03-9, a Causal Agent of Grapevine Crown Gall Disease. Molecular Plant-Microbe Interactions® 33, 1280-1282(2020). doi:10.1094/MPMI-07-20-0180-A
Yoshiteru Noutoshi, Atsushi Toyoda, Tomoya Ishii, Kirara Saito, Megumi Watanabe, Akira Kawaguchi. Complete Genome Sequence Data of Nonpathogenic and Nonantagonistic Strain of Rhizobium vitis VAR06-30 Isolated From Grapevine Rhizosphere. Molecular Plant-Microbe Interactions® 33, 1283-1285(2020). doi:10.1094/MPMI-07-20-0182-A
Hideo Matsumura, Min-Chien Hsiao, Ya-Ping Lin, Atsushi Toyoda, Naoki Taniai, Kazuhiko Tarora, Naoya Urasaki, Shashi S. Anand, Narinder P. S. Dhillon, Roland Schafleitner, Cheng-Ruei Lee. Reply to Renner: Meticulous investigation, not sequencing effort, leads to robust conclusion. Proceedings of the National Academy of Sciences 117, 24632-24633(2020). doi:10.1073/pnas.2015561117
Masato Nikaido, Shinji Kondo, Zicong Zhang, Jiaqi Wu, Hidenori Nishihara, Yoshihito Niimura, Shunta Suzuki, Kazushige Touhara, Yutaka Suzuki, Hideki Noguchi, Yohei Minakuchi, Atsushi Toyoda, Asao Fujiyama, Sumio Sugano, Misako Yoneda, Chieko Kai. Comparative genomic analyses illuminate the distinct evolution of megabats within Chiroptera. DNA Research 27, (2020). doi:10.1093/dnares/dsaa021
Nao Ikeyama, Takumi Murakami, Atsushi Toyoda, Hiroshi Mori, Takao Iino, Moriya Ohkuma, Mitsuo Sakamoto. Microbial interaction between the succinate‐utilizing bacterium Phascolarctobacterium faecium and the gut commensal Bacteroides thetaiotaomicron. MicrobiologyOpen 9, (2020). doi:10.1002/mbo3.1111
Takashi Gakuhari, Shigeki Nakagome, Simon Rasmussen, Morten E. Allentoft, Takehiro Sato, Thorfinn Korneliussen, Blánaid Ní Chuinneagáin, Hiromi Matsumae, Kae Koganebuchi, Ryan Schmidt, Souichiro Mizushima, Osamu Kondo, Nobuo Shigehara, Minoru Yoneda, Ryosuke Kimura, Hajime Ishida, Tadayuki Masuyama, Yasuhiro Yamada, Atsushi Tajima, Hiroki Shibata, Atsushi Toyoda, Toshiyuki Tsurumoto, Tetsuaki Wakebe, Hiromi Shitara, Tsunehiko Hanihara, Eske Willerslev, Martin Sikora, Hiroki Oota. Ancient Jomon genome sequence analysis sheds light on migration patterns of early East Asian populations. Communications Biology 3, (2020). doi:10.1038/s42003-020-01162-2
Naohisa Wada, Hideaki Yuasa, Rei Kajitani, Yasuhiro Gotoh, Yoshitoshi Ogura, Dai Yoshimura, Atsushi Toyoda, Sen-Lin Tang, Yukihiro Higashimura, Hugh Sweatman, Zac Forsman, Omri Bronstein, Gal Eyal, Nalinee Thongtham, Takehiko Itoh, Tetsuya Hayashi, Nina Yasuda. A ubiquitous subcuticular bacterial symbiont of a coral predator, the crown-of-thorns starfish, in the Indo-Pacific. Microbiome 8, (2020). doi:10.1186/s40168-020-00880-3
Hirokazu Yano, Md. Zobaidul Alam, Emiko Rimbara, Tomoko F. Shibata, Masaki Fukuyo, Yoshikazu Furuta, Tomoaki Nishiyama, Shuji Shigenobu, Mitsuyasu Hasebe, Atsushi Toyoda, Yutaka Suzuki, Sumio Sugano, Keigo Shibayama, Ichizo Kobayashi. Networking and Specificity-Changing DNA Methyltransferases in Helicobacter pylori. Frontiers in Microbiology 11, (2020). doi:10.3389/fmicb.2020.01628
Takashi Abe, Yu Akazawa, Atsushi Toyoda, Hironori Niki, Tomoya Baba. Batch-Learning Self-Organizing Map Identifies Horizontal Gene Transfer Candidates and Their Origins in Entire Genomes. Frontiers in Microbiology 11, (2020). doi:10.3389/fmicb.2020.01486
Kai Wang, Ryo Tomura, Wei Chen, Miho Kiyooka, Hinako Ishizaki, Tomoyuki Aizu, Yohei Minakuchi, Masahide Seki, Yutaka Suzuki, Tatsuya Omotezako, Ritsuko Suyama, Aki Masunaga, Charles Plessy, Nicholas M. Luscombe, Christelle Dantec, Patrick Lemaire, Takehiko Itoh, Atsushi Toyoda, Hiroki Nishida, Takeshi A. Onuma. A genome database for a Japanese population of the larvacean Oikopleura dioica. Development, Growth & Differentiation 62, 450-461(2020). doi:10.1111/dgd.12689
Yo Y. Yamasaki, Ryo Kakioka, Hiroshi Takahashi, Atsushi Toyoda, Atsushi J. Nagano, Yoshiyasu Machida, Peter R. Møller, Jun Kitano. Genome-wide patterns of divergence and introgression after secondary contact between Pungitius sticklebacks. Philosophical Transactions of the Royal Society B: Biological Sciences 375, 20190548(2020). doi:10.1098/rstb.2019.0548
Jun Kitano, Ryo Kakioka, Asano Ishikawa, Atsushi Toyoda, Makoto Kusakabe. Differences in the contributions of sex linkage and androgen regulation to sex‐biased gene expression in juvenile and adult sticklebacks. Journal of Evolutionary Biology 33, 1129-1138(2020). doi:10.1111/jeb.13662
Mariko Takeuchi, Hirokazu Kuwahara, Takumi Murakami, Kazuki Takahashi, Rei Kajitani, Atsushi Toyoda, Takehiko Itoh, Moriya Ohkuma, Yuichi Hongoh. Parallel reductive genome evolution in Desulfovibrio ectosymbionts independently acquired by Trichonympha protists in the termite gut. The ISME Journal 14, 2288-2301(2020). doi:10.1038/s41396-020-0688-1
Hideo Matsumura, Min-Chien Hsiao, Ya-Ping Lin, Atsushi Toyoda, Naoki Taniai, Kazuhiko Tarora, Naoya Urasaki, Shashi S. Anand, Narinder P. S. Dhillon, Roland Schafleitner, Cheng-Ruei Lee. Long-read bitter gourd (Momordica charantia) genome and the genomic architecture of nonclassic domestication. Proceedings of the National Academy of Sciences 117, 14543-14551(2020). doi:10.1073/pnas.1921016117
Nao Ikeyama, Atsushi Toyoda, Sho Morohoshi, Tadao Kunihiro, Takumi Murakami, Hiroshi Mori, Takao Iino, Moriya Ohkuma, Mitsuo Sakamoto. Amedibacterium intestinale gen. nov., sp. nov., isolated from human faeces, and reclassification of Eubacterium dolichum Moore et al. 1976 (Approved Lists 1980) as Amedibacillus dolichus gen. nov., comb. nov. International Journal of Systematic and Evolutionary Microbiology 70, 3656-3664(2020). doi:10.1099/ijsem.0.004215
Tetsuo Kon, Yoshihiro Omori, Kentaro Fukuta, Hironori Wada, Masakatsu Watanabe, Zelin Chen, Miki Iwasaki, Tappei Mishina, Shin-ichiro S. Matsuzaki, Daiki Yoshihara, Jumpei Arakawa, Koichi Kawakami, Atsushi Toyoda, Shawn M. Burgess, Hideki Noguchi, Takahisa Furukawa. The Genetic Basis of Morphological Diversity in Domesticated Goldfish. Current Biology 30, 2260-2274.e6(2020). doi:10.1016/j.cub.2020.04.034
Yuichi Fukutomi, Shu Kondo, Atsushi Toyoda, Shuji Shigenobu, Shigeyuki Koshikawa. Transcriptome analysis reveals wingless regulates neural development and signaling genes in the region of wing pigmentation of a polka‐dotted fruit fly. The FEBS Journal 288, 99-110(2020). doi:10.1111/febs.15338
Kohta Yoshida, Mark Ravinet, Takashi Makino, Atsushi Toyoda, Tomoyuki Kokita, Seiichi Mori, Jun Kitano. Accumulation of Deleterious Mutations in Landlocked Threespine Stickleback Populations. Genome Biology and Evolution 12, 479-492(2020). doi:10.1093/gbe/evaa065
Matt Shenton, Masaaki Kobayashi, Shin Terashima, Hajime Ohyanagi, Dario Copetti, Tania Hernández-Hernández, Jianwei Zhang, Nobuko Ohmido, Masahiro Fujita, Atsushi Toyoda, Hiroshi Ikawa, Asao Fujiyama, Hiroyasu Furuumi, Toshie Miyabayashi, Takahiko Kubo, David Kudrna, Rod Wing, Kentaro Yano, Ken-ichi Nonomura, Yutaka Sato, Nori Kurata. Evolution and diversity of the wild rice Oryza officinalis complex, across continents genome types, and ploidy levels. Genome Biology and Evolution , (2020). doi:10.1093/gbe/evaa037
Toshinori Hyodo, Md. Lutfur Rahman, Sivasundaram Karnan, Takuji Ito, Atsushi Toyoda, Akinobu Ota, Md Wahiduzzaman, Shinobu Tsuzuki, Yohei Okada, Yoshitaka Hosokawa, Hiroyuki Konishi. Tandem Paired Nicking Promotes Precise Genome Editing with Scarce Interference by p53. Cell Reports 30, 1195-1207.e7(2020). doi:10.1016/j.celrep.2019.12.064
Kenta Shirasawa, Hiroshi Yakushiji, Ryotaro Nishimura, Takeshige Morita, Shota Jikumaru, Hidetoshi Ikegami, Atsushi Toyoda, Hideki Hirakawa, Sachiko Isobe. The Ficus erecta genome aids Ceratocystis canker resistance breeding in common fig (F. carica). The Plant Journal 102, 1313-1322(2020). doi:10.1111/tpj.14703
Keiji Nakamura, Kazunori Murase, Mitsuhiko P. Sato, Atsushi Toyoda, Takehiko Itoh, Jacques Georges Mainil, Denis Piérard, Shuji Yoshino, Keiko Kimata, Junko Isobe, Kazuko Seto, Yoshiki Etoh, Hiroshi Narimatsu, Shioko Saito, Jun Yatsuyanagi, Kenichi Lee, Sunao Iyoda, Makoto Ohnishi, Tadasuke Ooka, Yasuhiro Gotoh, Yoshitoshi Ogura, Tetsuya Hayashi. Differential dynamics and impacts of prophages and plasmids on the pangenome and virulence factor repertoires of Shiga toxin-producing Escherichia coli O145:H28. Microbial Genomics 6, (2020). doi:10.1099/mgen.0.000323
Taro Tsujimura, Osamu Takase, Masahiro Yoshikawa, Etsuko Sano, Matsuhiko Hayashi, Kazuto Hoshi, Tsuyoshi Takato, Atsushi Toyoda, Hideyuki Okano, Keiichi Hishikawa. Controlling gene activation by enhancers through a drug-inducible topological insulator. eLife 9, (2020). doi:10.7554/eLife.47980
Natsune Takagaki, Akane Ohta, Kohei Ohnishi, Akira Kawanabe, Yohei Minakuchi, Atsushi Toyoda, Yuichiro Fujiwara, Atsushi Kuhara. The mechanoreceptor DEG‐1 regulates cold tolerance in Caenorhabditis elegans. EMBO reports 21, (2020). doi:10.15252/embr.201948671
Yasunari Satoh, Jun-ichi Asakawa, Mayumi Nishimura, Tony Kuo, Norio Shinkai, Harry M. Cullings, Yohei Minakuchi, Jun Sese, Atsushi Toyoda, Yoshiya Shimada, Nori Nakamura, Arikuni Uchimura. Characteristics of induced mutations in offspring derived from irradiated mouse spermatogonia and mature oocytes. Scientific Reports 10, (2020). doi:10.1038/s41598-019-56881-2
Mitsuo Sakamoto, Nao Ikeyama, Atsushi Toyoda, Takumi Murakami, Hiroshi Mori, Takao Iino, Moriya Ohkuma. Dialister hominis sp. nov., isolated from human faeces. International Journal of Systematic and Evolutionary Microbiology 70, 589-595(2020). doi:10.1099/ijsem.0.003797
Toshinori Hyodo, Md. Lutfur Rahman, Sivasundaram Karnan, Takuji Ito, Atsushi Toyoda, Akinobu Ota, Md Wahiduzzaman, Shinobu Tsuzuki, Yohei Okada, Yoshitaka Hosokawa, Hiroyuki Konishi. Tandem Paired Nicking Promotes Precise Genome Editing with Scarce Interference by p53. Cell Reports 30, 1195-1207.e7(2020). doi:10.1016/j.celrep.2019.12.064
- 2019年 -
Yu Zheng, Ayana Saitou, Chiung-Mei Wang, Atsushi Toyoda, Yohei Minakuchi, Yuji Sekiguchi, Kenji Ueda, Hideaki Takano, Yasuteru Sakai, Keietsu Abe, Akira Yokota, Shuhei Yabe. Genome Features and Secondary Metabolites Biosynthetic Potential of the Class Ktedonobacteria. Frontiers in Microbiology 10, (2019). doi:10.3389/fmicb.2019.00893
Shinichi Yachida, Sayaka Mizutani, Hirotsugu Shiroma, Satoshi Shiba, Takeshi Nakajima, Taku Sakamoto, Hikaru Watanabe, Keigo Masuda, Yuichiro Nishimoto, Masaru Kubo, Fumie Hosoda, Hirofumi Rokutan, Minori Matsumoto, Hiroyuki Takamaru, Masayoshi Yamada, Takahisa Matsuda, Motoki Iwasaki, Taiki Yamaji, Tatsuo Yachida, Tomoyoshi Soga, Ken Kurokawa, Atsushi Toyoda, Yoshitoshi Ogura, Tetsuya Hayashi, Masanori Hatakeyama, Hitoshi Nakagama, Yutaka Saito, Shinji Fukuda, Tatsuhiro Shibata, Takuji Yamada. Metagenomic and metabolomic analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer. Nature Medicine 25, 968-976(2019). doi:10.1038/s41591-019-0458-7
Chiung-mei Wang, Yu Zheng, Yasuteru Sakai, Atsushi Toyoda, Yohei Minakuchi, Keietsu Abe, Akira Yokota, Shuhei Yabe. Tengunoibacter tsumagoiensis gen. nov., sp. nov., Dictyobacter kobayashii sp. nov., Dictyobacter alpinus sp. nov., and description of Dictyobacteraceae fam. nov. within the order Ktedonobacterales isolated from Tengu-no-mugimeshi, a soil-like granular mass of micro-organisms, and emended descriptions of the genera Ktedonobacter and Dictyobacter. International Journal of Systematic and Evolutionary Microbiology 69, 1910-1918(2019). doi:10.1099/ijsem.0.003396
Nobu Sutra, Junko Kusumi, Javier Montenegro, Hirozumi Kobayashi, Shingo Fujimoto, Kawilarang W. A. Masengi, Atsushi J. Nagano, Atsushi Toyoda, Masatoshi Matsunami, Ryosuke Kimura, Kazunori Yamahira. Evidence for sympatric speciation in a Wallacean ancient lake. Evolution 73, 1898-1915(2019). doi:10.1111/evo.13821
Masatoshi Okura, Fumito Maruyama, Atsushi Ota, Takeshi Tanaka, Yohei Matoba, Aya Osawa, Sayed Mushtaq Sadaat, Makoto Osaki, Atsushi Toyoda, Yoshitoshi Ogura, Tetsuya Hayashi, Daisuke Takamatsu. Genotypic diversity of Streptococcus suis and the S. suis-like bacterium Streptococcus ruminantium in ruminants. Veterinary Research 50, (2019). doi:10.1186/s13567-019-0708-1
Michael T. Madigan, Sol M. Resnick, Megan L. Kempher, Alice C. Dohnalkova, Shinichi Takaichi, Zheng-Yu Wang-Otomo, Atsushi Toyoda, Ken Kurokawa, Hiroshi Mori, Yusuke Tsukatani. Blastochloris tepida, sp. nov., a thermophilic species of the bacteriochlorophyll b-containing genus Blastochloris. Archives of Microbiology 201, 1351-1359(2019). doi:10.1007/s00203-019-01701-4
Takashi Koyama, Masatoshi Nakamoto, Kagayaki Morishima, Ryohei Yamashita, Takefumi Yamashita, Kohei Sasaki, Yosuke Kuruma, Naoki Mizuno, Moe Suzuki, Yoshiharu Okada, Risa Ieda, Tsubasa Uchino, Satoshi Tasumi, Sho Hosoya, Seiichi Uno, Jiro Koyama, Atsushi Toyoda, Kiyoshi Kikuchi, Takashi Sakamoto. A SNP in a Steroidogenic Enzyme Is Associated with Phenotypic Sex in Seriola Fishes. Current Biology 29, 1901-1909.e8(2019). doi:10.1016/j.cub.2019.04.069
Yuta Kobayashi, Yosui Nojima, Takuma Sakamoto, Kikuo Iwabuchi, Takeru Nakazato, Hidemasa Bono, Atsushi Toyoda, Asao Fujiyama, Michael R. Kanost, Hiroko Tabunoki. Comparative analysis of seven types of superoxide dismutases for their ability to respond to oxidative stress in Bombyx mori. Scientific Reports 9, (2019). doi:10.1038/s41598-018-38384-8
Tomohiko Kazama, Miki Okuno, Yuta Watari, Shungo Yanase, Chie Koizuka, Yu Tsuruta, Hajime Sugaya, Atsushi Toyoda, Takehiko Itoh, Nobuhiro Tsutsumi, Kinya Toriyama, Nobuya Koizuka, Shin-ichi Arimura. Curing cytoplasmic male sterility via TALEN-mediated mitochondrial genome editing. Nature Plants 5, 722-730(2019). doi:10.1038/s41477-019-0459-z
Munetaka Kawamoto, Akiya Jouraku, Atsushi Toyoda, Kakeru Yokoi, Yohei Minakuchi, Susumu Katsuma, Asao Fujiyama, Takashi Kiuchi, Kimiko Yamamoto, Toru Shimada. High-quality genome assembly of the silkworm, Bombyx mori. Insect Biochemistry and Molecular Biology 107, 53-62(2019). doi:10.1016/j.ibmb.2019.02.002
Rei Kajitani, Dai Yoshimura, Miki Okuno, Yohei Minakuchi, Hiroshi Kagoshima, Asao Fujiyama, Kaoru Kubokawa, Yuji Kohara, Atsushi Toyoda, Takehiko Itoh. Platanus-allee is a de novo haplotype assembler enabling a comprehensive access to divergent heterozygous regions. Nature Communications 10, (2019). doi:10.1038/s41467-019-09575-2
Ito, Nishio, Tarutani, Emura, Honjo, Toyoda, Fujiyama, Kakutani, Kudoh. Seasonal Stability and Dynamics of DNA Methylation in Plants in a Natural Environment. Genes 10, 544(2019). doi:10.3390/genes10070544
Kiyoe Ishimoto, Shino Sohonahra, Mitsuko Kishi-Kaboshi, Jun-ichi Itoh, Ken-ichiro Hibara, Yutaka Sato, Tsuneaki Watanabe, Kiyomi Abe, Akio Miyao, Misuzu Nosaka-Takahashi, Toshiya Suzuki, Nhung Kim Ta, Sae Shimizu-Sato, Takamasa Suzuki, Atsushi Toyoda, Hirokazu Takahashi, Mikio Nakazono, Yasuo Nagato, Hirohiko Hirochika, Yutaka Sato. Specification of basal region identity after asymmetric zygotic division requires mitogen-activated protein kinase 6 in rice. Development 146, dev176305(2019). doi:10.1242/dev.176305
Asano Ishikawa, Naoki Kabeya, Koki Ikeya, Ryo Kakioka, Jennifer N. Cech, Naoki Osada, Miguel C. Leal, Jun Inoue, Manabu Kume, Atsushi Toyoda, Ayumi Tezuka, Atsushi J. Nagano, Yo Y. Yamasaki, Yuto Suzuki, Tomoyuki Kokita, Hiroshi Takahashi, Kay Lucek, David Marques, Yusuke Takehana, Kiyoshi Naruse, Seiichi Mori, Oscar Monroig, Nemiah Ladd, Carsten J. Schubert, Blake Matthews, Catherine L. Peichel, Ole Seehausen, Goro Yoshizaki, Jun Kitano. A key metabolic gene for recurrent freshwater colonization and radiation in fishes. Science 364, 886-889(2019). doi:10.1126/science.aau5656
Satoshi Hiraoka, Yusuke Okazaki, Mizue Anda, Atsushi Toyoda, Shin-ichi Nakano, Wataru Iwasaki. Metaepigenomic analysis reveals the unexplored diversity of DNA methylation in an environmental prokaryotic community. Nature Communications 10, (2019). doi:10.1038/s41467-018-08103-y
Omaya Dudin, Andrej Ondracka, Xavier Grau-Bové, Arthur AB Haraldsen, Atsushi Toyoda, Hiroshi Suga, Jon Bråte, Iñaki Ruiz-Trillo. A unicellular relative of animals generates a layer of polarized cells by actomyosin-dependent cellularization. eLife 8, (2019). doi:10.7554/eLife.49801
Zelin Chen, Yoshihiro Omori, Sergey Koren, Takuya Shirokiya, Takuo Kuroda, Atsushi Miyamoto, Hironori Wada, Asao Fujiyama, Atsushi Toyoda, Suiyuan Zhang, Tyra G. Wolfsberg, Koichi Kawakami, Adam M. Phillippy, James C. Mullikin, Shawn M. Burgess. De novo assembly of the goldfish (Carassius auratus) genome and the evolution of genes after whole-genome duplication. Science Advances 5, eaav0547(2019). doi:10.1126/sciadv.aav0547
- 2018年 -
Kohta Yoshida, Asano Ishikawa, Atsushi Toyoda, Shuji Shigenobu, Asao Fujiyama, Jun Kitano. Functional divergence of a heterochromatin‐binding protein during stickleback speciation. Molecular Ecology 28, 1563-1578(2018). doi:10.1111/mec.14841
Tomoyo Ujisawa, Akane Ohta, Tatsuya Ii, Yohei Minakuchi, Atsushi Toyoda, Miki Ii, Atsushi Kuhara. Endoribonuclease ENDU-2 regulates multiple traits including cold tolerance via cell autonomous and nonautonomous controls in Caenorhabditis elegans. Proceedings of the National Academy of Sciences 115, 8823-8828(2018). doi:10.1073/pnas.1808634115
Taro Tsujimura, Osamu Takase, Masahiro Yoshikawa, Etsuko Sano, Matsuhiko Hayashi, Tsuyoshi Takato, Atsushi Toyoda, Hideyuki Okano, Keiichi Hishikawa. Control of directionality of chromatin folding for the inter- and intra-domain contacts at the Tfap2c–Bmp7 locus. Epigenetics & Chromatin 11, (2018). doi:10.1186/s13072-018-0221-1
Tomohiro Tsuji, Hiroki Gotoh, Shinichi Morita, Junya Hirata, Yohei Minakuchi, Toshinobu Yaginuma, Atsushi Toyoda, Teruyuki Niimi. Molecular Characterization of Eye Pigmentation-Related ABC Transporter Genes in the Ladybird Beetle Harmonia axyridis Reveals Striking Gene Duplication of the white Gene. Zoological Science 35, 260(2018). doi:10.2108/zs170166
Yasuhiro Tanizawa, Ipputa Tada, Hisami Kobayashi, Akihito Endo, Shintaro Maeno, Atsushi Toyoda, Masanori Arita, Yasukazu Nakamura, Mitsuo Sakamoto, Moriya Ohkuma, Masanori Tohno. Lactobacillus paragasseri sp. nov., a sister taxon of Lactobacillus gasseri, based on whole-genome sequence analyses. International Journal of Systematic and Evolutionary Microbiology 68, 3512-3517(2018). doi:10.1099/ijsem.0.003020
Stefan Reuscher, Tomoyuki Furuta, Kanako Bessho-Uehara, Michele Cosi, Kshirod K. Jena, Atsushi Toyoda, Asao Fujiyama, Nori Kurata, Motoyuki Ashikari. Assembling the genome of the African wild rice Oryza longistaminata by exploiting synteny in closely related Oryza species. Communications Biology 1, (2018). doi:10.1038/s42003-018-0171-y
Mark Ravinet, Kohta Yoshida, Shuji Shigenobu, Atsushi Toyoda, Asao Fujiyama, Jun Kitano. The genomic landscape at a late stage of stickleback speciation: High genomic divergence interspersed by small localized regions of introgression. PLOS Genetics 14, e1007358(2018). doi:10.1371/journal.pgen.1007358
Yoshitoshi Ogura, Kazuko Seto, Yo Morimoto, Keiji Nakamura, Mitsuhiko P. Sato, Yasuhiro Gotoh, Takehiko Itoh, Atsushi Toyoda, Makoto Ohnishi, Tetsuya Hayashi. Genomic Characterization of β-Glucuronidase–Positive Escherichia coli O157:H7 Producing Stx2a. Emerging Infectious Diseases 24, 2219-2227(2018). doi:10.3201/eid2412.180404
Tomoaki Nishiyama, Hidetoshi Sakayama, Jan de Vries, Henrik Buschmann, Denis Saint-Marcoux, Kristian K. Ullrich, Fabian B. Haas, Lisa Vanderstraeten, Dirk Becker, Daniel Lang, Stanislav Vosolsobě, Stephane Rombauts, Per K.I. Wilhelmsson, Philipp Janitza, Ramona Kern, Alexander Heyl, Florian Rümpler, Luz Irina A. Calderón Villalobos, John M. Clay, Roman Skokan, Atsushi Toyoda, Yutaka Suzuki, Hiroshi Kagoshima, Elio Schijlen, Navindra Tajeshwar, Bruno Catarino, Alexander J. Hetherington, Assia Saltykova, Clemence Bonnot, Holger Breuninger, Aikaterini Symeonidi, Guru V. Radhakrishnan, Filip Van Nieuwerburgh, Dieter Deforce, Caren Chang, Kenneth G. Karol, Rainer Hedrich, Peter Ulvskov, Gernot Glöckner, Charles F. Delwiche, Jan Petrášek, Yves Van de Peer, Jiri Friml, Mary Beilby, Liam Dolan, Yuji Kohara, Sumio Sugano, Asao Fujiyama, Pierre-Marc Delaux, Marcel Quint, Günter Theißen, Martin Hagemann, Jesper Harholt, Christophe Dunand, Sabine Zachgo, Jane Langdale, Florian Maumus, Dominique Van Der Straeten, Sven B. Gould, Stefan A. Rensing. The Chara Genome: Secondary Complexity and Implications for Plant Terrestrialization. Cell 174, 448-464.e24(2018). doi:10.1016/j.cell.2018.06.033
Kazue Nakanaga, Yoshitoshi Ogura, Atsushi Toyoda, Mitsunori Yoshida, Hanako Fukano, Nagatoshi Fujiwara, Yuji Miyamoto, Noboru Nakata, Yuko Kazumi, Shinji Maeda, Tadasuke Ooka, Masamichi Goto, Kazunari Tanigawa, Satoshi Mitarai, Koichi Suzuki, Norihisa Ishii, Manabu Ato, Tetsuya Hayashi, Yoshihiko Hoshino. Naturally occurring a loss of a giant plasmid from Mycobacterium ulcerans subsp. shinshuense makes it non-pathogenic. Scientific Reports 8, (2018). doi:10.1038/s41598-018-26425-1
Anzu Minami, Kenji Yano, Rico Gamuyao, Keisuke Nagai, Takeshi Kuroha, Madoka Ayano, Masanari Nakamori, Masaya Koike, Yuma Kondo, Yoko Niimi, Keiko Kuwata, Takamasa Suzuki, Tetsuya Higashiyama, Yumiko Takebayashi, Mikiko Kojima, Hitoshi Sakakibara, Atsushi Toyoda, Asao Fujiyama, Nori Kurata, Motoyuki Ashikari, Stefan Reuscher. Time-Course Transcriptomics Analysis Reveals Key Responses of Submerged Deepwater Rice to Flooding. Plant Physiology 176, 3081-3102(2018). doi:10.1104/pp.17.00858
Hugh McColl, Fernando Racimo, Lasse Vinner, Fabrice Demeter, Takashi Gakuhari, J. Víctor Moreno-Mayar, George van Driem, Uffe Gram Wilken, Andaine Seguin-Orlando, Constanza de la Fuente Castro, Sally Wasef, Rasmi Shoocongdej, Viengkeo Souksavatdy, Thongsa Sayavongkhamdy, Mohd Mokhtar Saidin, Morten E. Allentoft, Takehiro Sato, Anna-Sapfo Malaspinas, Farhang A. Aghakhanian, Thorfinn Korneliussen, Ana Prohaska, Ashot Margaryan, Peter de Barros Damgaard, Supannee Kaewsutthi, Patcharee Lertrit, Thi Mai Huong Nguyen, Hsiao-chun Hung, Thi Minh Tran, Huu Nghia Truong, Giang Hai Nguyen, Shaiful Shahidan, Ketut Wiradnyana, Hiromi Matsumae, Nobuo Shigehara, Minoru Yoneda, Hajime Ishida, Tadayuki Masuyama, Yasuhiro Yamada, Atsushi Tajima, Hiroki Shibata, Atsushi Toyoda, Tsunehiko Hanihara, Shigeki Nakagome, Thibaut Deviese, Anne-Marie Bacon, Philippe Duringer, Jean-Luc Ponche, Laura Shackelford, Elise Patole-Edoumba, Anh Tuan Nguyen, Bérénice Bellina-Pryce, Jean-Christophe Galipaud, Rebecca Kinaston, Hallie Buckley, Christophe Pottier, Simon Rasmussen, Tom Higham, Robert A. Foley, Marta Mirazón Lahr, Ludovic Orlando, Martin Sikora, Maude E. Phipps, Hiroki Oota, Charles Higham, David M. Lambert, Eske Willerslev. The prehistoric peopling of Southeast Asia. Science 361, 88-92(2018). doi:10.1126/science.aat3628
Fumika Ichino, Hidemasa Bono, Takeru Nakazato, Atsushi Toyoda, Asao Fujiyama, Kikuo Iwabuchi, Ryoichi Sato, Hiroko Tabunoki. Construction of a simple evaluation system for the intestinal absorption of an orally administered medicine using Bombyx mori larvae. Drug Discoveries & Therapeutics 12, 7-15(2018). doi:10.5582/ddt.2018.01004
Takashi Hamaji, Hiroko Kawai-Toyooka, Haruka Uchimura, Masahiro Suzuki, Hideki Noguchi, Yohei Minakuchi, Atsushi Toyoda, Asao Fujiyama, Shin-ya Miyagishima, James G. Umen, Hisayoshi Nozaki. Anisogamy evolved with a reduced sex-determining region in volvocine green algae. Communications Biology 1, (2018). doi:10.1038/s42003-018-0019-5
Hager Gaouda, Takashi Hamaji, Kayoko Yamamoto, Hiroko Kawai-Toyooka, Masahiro Suzuki, Hideki Noguchi, Yohei Minakuchi, Atsushi Toyoda, Asao Fujiyama, Hisayoshi Nozaki, David Roy Smith. Exploring the Limits and Causes of Plastid Genome Expansion in Volvocine Green Algae. Genome Biology and Evolution 10, 2248-2254(2018). doi:10.1093/gbe/evy175
Yu Chen, Yoko Kuroki, Geoff Shaw, Andrew Pask, Hongshi Yu, Atsushi Toyoda, Asao Fujiyama, Marilyn Renfree. Androgen and Oestrogen Affect the Expression of Long Non-Coding RNAs During Phallus Development in a Marsupial. Non-Coding RNA 5, 3(2018). doi:10.3390/ncrna5010003
Toshiya Ando, Takeshi Matsuda, Kumiko Goto, Kimiko Hara, Akinori Ito, Junya Hirata, Joichiro Yatomi, Rei Kajitani, Miki Okuno, Katsushi Yamaguchi, Masaaki Kobayashi, Tomoyuki Takano, Yohei Minakuchi, Masahide Seki, Yutaka Suzuki, Kentaro Yano, Takehiko Itoh, Shuji Shigenobu, Atsushi Toyoda, Teruyuki Niimi. Repeated inversions within a pannier intron drive diversification of intraspecific colour patterns of ladybird beetles. Nature Communications 9, (2018). doi:10.1038/s41467-018-06116-1
Yang An, Akane Kawaguchi, Chen Zhao, Atsushi Toyoda, Ali Sharifi-Zarchi, Seyed Ahmad Mousavi, Reza Bagherzadeh, Takeshi Inoue, Hajime Ogino, Asao Fujiyama, Hamidreza Chitsaz, Hossein Baharvand, Kiyokazu Agata. Draft genome of Dugesia japonica provides insights into conserved regulatory elements of the brain restriction gene nou-darake in planarians. Zoological Letters 4, (2018). doi:10.1186/s40851-018-0102-2
- 2017年 -
Kousuke Mouri, Tomoko Sagai, Akiteru Maeno, Takanori Amano, Atsushi Toyoda, Toshihiko Shiroishi. Enhancer adoption caused by genomic insertion elicits interdigital Shh expression and syndactyly in mouse. Proceedings of the National Academy of Sciences 115, 1021-1026(2017). doi:10.1073/pnas.1713339115
Tomohiro Kudoh, Mitsuhiko Takahashi, Takayuki Osabe, Atsushi Toyoda, Hideki Hirakawa, Yutaka Suzuki, Nobuko Ohmido, Yasuyuki Onodera. Molecular insights into the non-recombining nature of the spinach male-determining region. Molecular Genetics and Genomics 293, 557-568(2017). doi:10.1007/s00438-017-1405-2
Kentaro Hori, Yasuyuki Yamada, Ratmoyo Purwanto, Yohei Minakuchi, Atsushi Toyoda, Hideki Hirakawa, Fumihiko Sato. Mining of the Uncharacterized Cytochrome P450 Genes Involved in Alkaloid Biosynthesis in California Poppy Using a Draft Genome Sequence. Plant and Cell Physiology 59, 222-233(2017). doi:10.1093/pcp/pcx210
Tomokazu Yamazaki, Kensuke Ichihara, Ryogo Suzuki, Kenshiro Oshima, Shinichi Miyamura, Kazuyoshi Kuwano, Atsushi Toyoda, Yutaka Suzuki, Sumio Sugano, Masahira Hattori, Shigeyuki Kawano. Genomic structure and evolution of the mating type locus in the green seaweed Ulva partita. Scientific Reports 7, (2017). doi:10.1038/s41598-017-11677-0
Sayaka Tsuchida, Fumito Maruyama, Yoshitoshi Ogura, Atsushi Toyoda, Tetsuya Hayashi, Moriya Okuma, Kazunari Ushida. Genomic Characteristics of Bifidobacterium thermacidophilum Pig Isolates and Wild Boar Isolates Reveal the Unique Presence of a Putative Mobile Genetic Element with tetW for Pig Farm Isolates. Frontiers in Microbiology 8, (2017). doi:10.3389/fmicb.2017.01540
Hideto Takami, Atsushi Toyoda, Ikuo Uchiyama, Takehiko Itoh, Yoshihiro Takaki, Wataru Arai, Shinro Nishi, Mikihiko Kawai, Kazuo Shin-ya, Haruo Ikeda. Complete genome sequence and expression profile of the commercial lytic enzyme producerLysobacter enzymogenesM497-1. DNA Research , dsw055(2017). doi:10.1093/dnares/dsw055
Tokurou Shimizu, Yasuhiro Tanizawa, Takako Mochizuki, Hideki Nagasaki, Terutaka Yoshioka, Atsushi Toyoda, Asao Fujiyama, Eli Kaminuma, Yasukazu Nakamura. Draft Sequencing of the Heterozygous Diploid Genome of Satsuma (Citrus unshiu Marc.) Using a Hybrid Assembly Approach. Frontiers in Genetics 8, (2017). doi:10.3389/fgene.2017.00180
Ajeng K. Pramono, Hirokazu Kuwahara, Takehiko Itoh, Atsushi Toyoda, Akinori Yamada, Yuichi Hongoh. Discovery and Complete Genome Sequence of a Bacteriophage from an Obligate Intracellular Symbiont of a Cellulolytic Protist in the Termite Gut. Microbes and Environments 32, 112-117(2017). doi:10.1264/jsme2.ME16175
Takako Mochizuki, Yasuhiro Tanizawa, Takatomo Fujisawa, Tazro Ohta, Naruo Nikoh, Tokurou Shimizu, Atsushi Toyoda, Asao Fujiyama, Nori Kurata, Hideki Nagasaki, Eli Kaminuma, Yasukazu Nakamura. DNApod: DNA polymorphism annotation database from next-generation sequence read archives. PLOS ONE 12, e0172269(2017). doi:10.1371/journal.pone.0172269
Mai F. Minamikawa, Keisuke Nonaka, Eli Kaminuma, Hiromi Kajiya-Kanegae, Akio Onogi, Shingo Goto, Terutaka Yoshioka, Atsushi Imai, Hiroko Hamada, Takeshi Hayashi, Satomi Matsumoto, Yuichi Katayose, Atsushi Toyoda, Asao Fujiyama, Yasukazu Nakamura, Tokurou Shimizu, Hiroyoshi Iwata. Genome-wide association study and genomic prediction in citrus: Potential of genomics-assisted breeding for fruit quality traits. Scientific Reports 7, (2017). doi:10.1038/s41598-017-05100-x
Akira Kikuchi, Takeru Nakazato, Katsuhiko Ito, Yosui Nojima, Takeshi Yokoyama, Kikuo Iwabuchi, Hidemasa Bono, Atsushi Toyoda, Asao Fujiyama, Ryoichi Sato, Hiroko Tabunoki. Identification of functional enolase genes of the silkworm Bombyx mori from public databases with a combination of dry and wet bench processes. BMC Genomics 18, (2017). doi:10.1186/s12864-016-3455-y
Naho Kanda, Machiko Ichikawa, Ayaka Ono, Atsushi Toyoda, Asao Fujiyama, Jun Abe, Yuki Tsuchikane, Tomoaki Nishiyama, Hiroyuki Sekimoto. CRISPR/Cas9-based knockouts reveal that CpRLP1 is a negative regulator of the sex pheromone PR-IP in the Closterium peracerosum-strigosum-littorale complex. Scientific Reports 7, (2017). doi:10.1038/s41598-017-18251-8
Masafumi Kadowaki, Yuki Fujimaru, Seiga Taguchi, Jannatul Ferdouse, Kazutaka Sawada, Yuta Kimura, Yohei Terasawa, Gennaro Agrimi, Toyoaki Anai, Hideki Noguchi, Atsushi Toyoda, Asao Fujiyama, Takeshi Akao, Hiroshi Kitagaki. Chromosomal Aneuploidy Improves the Brewing Characteristics of Sake Yeast. Applied and Environmental Microbiology 83, (2017). doi:10.1128/AEM.01620-17
Asano Ishikawa, Makoto Kusakabe, Kohta Yoshida, Mark Ravinet, Takashi Makino, Atsushi Toyoda, Asao Fujiyama, Jun Kitano. Different contributions of local- and distant-regulatory changes to transcriptome divergence between stickleback ecotypes. Evolution 71, 565-581(2017). doi:10.1111/evo.13175
Soichi Inagaki, Mayumi Takahashi, Aoi Hosaka, Tasuku Ito, Atsushi Toyoda, Asao Fujiyama, Yoshiaki Tarutani, Tetsuji Kakutani. Gene‐body chromatin modification dynamics mediate epigenome differentiation in Arabidopsis. The EMBO Journal 36, 970-980(2017). doi:10.15252/embj.201694983
Aoi Hosaka, Raku Saito, Kazuya Takashima, Taku Sasaki, Yu Fu, Akira Kawabe, Tasuku Ito, Atsushi Toyoda, Asao Fujiyama, Yoshiaki Tarutani, Tetsuji Kakutani. Evolution of sequence-specific anti-silencing systems in Arabidopsis. Nature Communications 8, (2017). doi:10.1038/s41467-017-02150-7
Masahito Hayatsu, Kanako Tago, Ikuo Uchiyama, Atsushi Toyoda, Yong Wang, Yumi Shimomura, Takashi Okubo, Futoshi Kurisu, Yuhei Hirono, Kunihiko Nonaka, Hiroko Akiyama, Takehiko Itoh, Hideto Takami. An acid-tolerant ammonia-oxidizing γ-proteobacterium from soil. The ISME Journal 11, 1130-1141(2017). doi:10.1038/ismej.2016.191
Ritsuko Arai, Takeshi Sugawara, Yuko Sato, Yohei Minakuchi, Atsushi Toyoda, Kentaro Nabeshima, Hiroshi Kimura, Akatsuki Kimura. Reduction in chromosome mobility accompanies nuclear organization during early embryogenesis in Caenorhabditis elegans. Scientific Reports 7, (2017). doi:10.1038/s41598-017-03483-5
Arzuba Akter, Tadasuke Ooka, Yasuhiro Gotoh, Seigo Yamamoto, Hiromi Fujita, Fumio Terasoma, Kouji Kida, Masakatsu Taira, Fumiko Nakadouzono, Mutsuyo Gokuden, Manabu Hirano, Mamoru Miyashiro, Kouichi Inari, Yukie Shimazu, Kenji Tabara, Atsushi Toyoda, Dai Yoshimura, Takehiko Itoh, Tomokazu Kitano, Mitsuhiko P. Sato, Keisuke Katsura, Shakhinur Islam Mondal, Yoshitoshi Ogura, Shuji Ando, Tetsuya Hayashi. Extremely low genomic diversity of Rickettsia japonica distributed in Japan. Genome Biology and Evolution , evw304(2017). doi:10.1093/gbe/evw304
- 2016年 -
Makoto Kusakabe, Asano Ishikawa, Mark Ravinet, Kohta Yoshida, Takashi Makino, Atsushi Toyoda, Asao Fujiyama, Jun Kitano. Genetic basis for variation in salinity tolerance between stickleback ecotypes. Molecular Ecology 26, 304-319(2016). doi:10.1111/mec.13875
Tetsuya Hori, Naoko Kagawa, Atsushi Toyoda, Asao Fujiyama, Sadahiko Misu, Norikazu Monma, Fumiaki Makino, Kazuho Ikeo, Tatsuo Fukagawa. Constitutive centromere-associated network controls centromere drift in vertebrate cells. Journal of Cell Biology 216, 101-113(2016). doi:10.1083/jcb.201605001
Mitsunori Yoshida, Kazue Nakanaga, Yoshitoshi Ogura, Atsushi Toyoda, Tadasuke Ooka, Yuko Kazumi, Satoshi Mitarai, Norihisa Ishii, Tetsuya Hayashi, Yoshihiko Hoshino. Complete Genome Sequence of Mycobacterium ulcerans subsp. shinshuense. Genome Announcements 4, (2016). doi:10.1128/genomeA.01050-16
Kohta Yoshida, Ryutaro Miyagi, Seiichi Mori, Aya Takahashi, Takashi Makino, Atsushi Toyoda, Asao Fujiyama, Jun Kitano. Whole-genome sequencing reveals small genomic regions of introgression in an introduced crater lake population of threespine stickleback. Ecology and Evolution 6, 2190-2204(2016). doi:10.1002/ece3.2047
Kyoko Yoshida, Yasuhiro Go, Itaru Kushima, Atsushi Toyoda, Asao Fujiyama, Hiroo Imai, Nobuhito Saito, Atsushi Iriki, Norio Ozaki, Masaki Isoda. Single-neuron and genetic correlates of autistic behavior in macaque. Science Advances 2, e1600558(2016). doi:10.1126/sciadv.1600558
Tokurou Shimizu, Akira Kitajima, Keisuke Nonaka, Terutaka Yoshioka, Satoshi Ohta, Shingo Goto, Atsushi Toyoda, Asao Fujiyama, Takako Mochizuki, Hideki Nagasaki, Eli Kaminuma, Yasukazu Nakamura. Hybrid Origins of Citrus Varieties Inferred from DNA Marker Analysis of Nuclear and Organelle Genomes. PLOS ONE 11, e0166969(2016). doi:10.1371/journal.pone.0166969
Wei-Hao Shang, Tetsuya Hori, Frederick G. Westhorpe, Kristina M. Godek, Atsushi Toyoda, Sadahiko Misu, Norikazu Monma, Kazuho Ikeo, Christopher W. Carroll, Yasunari Takami, Asao Fujiyama, Hiroshi Kimura, Aaron F. Straight, Tatsuo Fukagawa. Acetylation of histone H4 lysine 5 and 12 is required for CENP-A deposition into centromeres. Nature Communications 7, (2016). doi:10.1038/ncomms13465
Adam M. Session, Yoshinobu Uno, Taejoon Kwon, Jarrod A. Chapman, Atsushi Toyoda, Shuji Takahashi, Akimasa Fukui, Akira Hikosaka, Atsushi Suzuki, Mariko Kondo, Simon J. van Heeringen, Ian Quigley, Sven Heinz, Hajime Ogino, Haruki Ochi, Uffe Hellsten, Jessica B. Lyons, Oleg Simakov, Nicholas Putnam, Jonathan Stites, Yoko Kuroki, Toshiaki Tanaka, Tatsuo Michiue, Minoru Watanabe, Ozren Bogdanovic, Ryan Lister, Georgios Georgiou, Sarita S. Paranjpe, Ila van Kruijsbergen, Shengquiang Shu, Joseph Carlson, Tsutomu Kinoshita, Yuko Ohta, Shuuji Mawaribuchi, Jerry Jenkins, Jane Grimwood, Jeremy Schmutz, Therese Mitros, Sahar V. Mozaffari, Yutaka Suzuki, Yoshikazu Haramoto, Takamasa S. Yamamoto, Chiyo Takagi, Rebecca Heald, Kelly Miller, Christian Haudenschild, Jacob Kitzman, Takuya Nakayama, Yumi Izutsu, Jacques Robert, Joshua Fortriede, Kevin Burns, Vaneet Lotay, Kamran Karimi, Yuuri Yasuoka, Darwin S. Dichmann, Martin F. Flajnik, Douglas W. Houston, Jay Shendure, Louis DuPasquier, Peter D. Vize, Aaron M. Zorn, Michihiko Ito, Edward M. Marcotte, John B. Wallingford, Yuzuru Ito, Makoto Asashima, Naoto Ueno, Yoichi Matsuda, Gert Jan C. Veenstra, Asao Fujiyama, Richard M. Harland, Masanori Taira, Daniel S. Rokhsar. Genome evolution in the allotetraploid frog Xenopus laevis. Nature 538, 336-343(2016). doi:10.1038/nature19840
Misaki Okahata, Akane Ohta, Hitomi Mizutani, Yohei Minakuchi, Atsushi Toyoda, Atsushi Kuhara. Natural variations of cold tolerance and temperature acclimation in Caenorhabditis elegans. Journal of Comparative Physiology B 186, 985-998(2016). doi:10.1007/s00360-016-1011-3
Ryosuke Nakai, Takatomo Fujisawa, Yasukazu Nakamura, Hiroyo Nishide, Ikuo Uchiyama, Tomoya Baba, Atsushi Toyoda, Asao Fujiyama, Takeshi Naganuma, Hironori Niki. Complete Genome Sequence of Aurantimicrobium minutum Type Strain KNCT, a Planktonic Ultramicrobacterium Isolated from River Water. Genome Announcements 4, (2016). doi:10.1128/genomeA.00616-16
Atsushi Hoshino, Vasanthan Jayakumar, Eiji Nitasaka, Atsushi Toyoda, Hideki Noguchi, Takehiko Itoh, Tadasu Shin-I, Yohei Minakuchi, Yuki Koda, Atsushi J. Nagano, Masaki Yasugi, Mie N. Honjo, Hiroshi Kudoh, Motoaki Seki, Asako Kamiya, Toshiyuki Shiraki, Piero Carninci, Erika Asamizu, Hiroyo Nishide, Sachiko Tanaka, Kyeung-Il Park, Yasumasa Morita, Kohei Yokoyama, Ikuo Uchiyama, Yoshikazu Tanaka, Satoshi Tabata, Kazuo Shinozaki, Yoshihide Hayashizaki, Yuji Kohara, Yutaka Suzuki, Sumio Sugano, Asao Fujiyama, Shigeru Iida, Yasubumi Sakakibara. Genome sequence and analysis of the Japanese morning glory Ipomoea nil. Nature Communications 7, (2016). doi:10.1038/ncomms13295
Takuma Hashimoto, Daiki D. Horikawa, Yuki Saito, Hirokazu Kuwahara, Hiroko Kozuka-Hata, Tadasu Shin-I, Yohei Minakuchi, Kazuko Ohishi, Ayuko Motoyama, Tomoyuki Aizu, Atsushi Enomoto, Koyuki Kondo, Sae Tanaka, Yuichiro Hara, Shigeyuki Koshikawa, Hiroshi Sagara, Toru Miura, Shin-ichi Yokobori, Kiyoshi Miyagawa, Yutaka Suzuki, Takeo Kubo, Masaaki Oyama, Yuji Kohara, Asao Fujiyama, Kazuharu Arakawa, Toshiaki Katayama, Atsushi Toyoda, Takekazu Kunieda. Extremotolerant tardigrade genome and improved radiotolerance of human cultured cells by tardigrade-unique protein. Nature Communications 7, (2016). doi:10.1038/ncomms12808
Erik R. Hanschen, Tara N. Marriage, Patrick J. Ferris, Takashi Hamaji, Atsushi Toyoda, Asao Fujiyama, Rafik Neme, Hideki Noguchi, Yohei Minakuchi, Masahiro Suzuki, Hiroko Kawai-Toyooka, David R. Smith, Halle Sparks, Jaden Anderson, Robert Bakarić, Victor Luria, Amir Karger, Marc W. Kirschner, Pierre M. Durand, Richard E. Michod, Hisayoshi Nozaki, Bradley J. S. C. Olson. The Gonium pectorale genome demonstrates co-option of cell cycle regulation during the evolution of multicellularity. Nature Communications 7, (2016). doi:10.1038/ncomms11370
Takashi Hamaji, Yuko Mogi, Patrick J. Ferris, Toshiyuki Mori, Shinya Miyagishima, Yukihiro Kabeya, Yoshiki Nishimura, Atsushi Toyoda, Hideki Noguchi, Asao Fujiyama, Bradley J. S. C. Olson, Tara N. Marriage, Ichiro Nishii, James G. Umen, Hisayoshi Nozaki. Sequence of theGonium pectoraleMating Locus Reveals a Complex and Dynamic History of Changes in Volvocine Algal Mating Haplotypes. G3: Genes, Genomes, Genetics 6, 1179-1189(2016). doi:10.1534/g3.115.026229
- 2015年 -
Kazunari Ushida, Sayaka Tsuchida, Yoshitoshi Ogura, Atsushi Toyoda, Fumito Maruyama. Domestication and cereal feeding developed domestic pig-type intestinal microbiota in animals of suidae. Animal Science Journal 87, 835-841(2015). doi:10.1111/asj.12492
Hajime Ohyanagi, Toshinobu Ebata, Xuehui Huang, Hao Gong, Masahiro Fujita, Takako Mochizuki, Atsushi Toyoda, Asao Fujiyama, Eli Kaminuma, Yasukazu Nakamura, Qi Feng, Zi-Xuan Wang, Bin Han, Nori Kurata. OryzaGenome: Genome Diversity Database of WildOryzaSpecies. Plant and Cell Physiology 57, e1-e1(2015). doi:10.1093/pcp/pcv171
Hideko Urushihara, Hidekazu Kuwayama, Kensuke Fukuhara, Takehiko Itoh, Hiroshi Kagoshima, Tadasu Shin-I, Atsushi Toyoda, Kazuyo Ohishi, Tateaki Taniguchi, Hideki Noguchi, Yoko Kuroki, Takashi Hata, Kyoko Uchi, Kurato Mohri, Jason S King, Robert H Insall, Yuji Kohara, Asao Fujiyama. Comparative genome and transcriptome analyses of the social amoeba Acytostelium subglobosum that accomplishes multicellular development without germ-soma differentiation. BMC Genomics 16, (2015). doi:10.1186/s12864-015-1278-x
Arikuni Uchimura, Mayumi Higuchi, Yohei Minakuchi, Mizuki Ohno, Atsushi Toyoda, Asao Fujiyama, Ikuo Miura, Shigeharu Wakana, Jo Nishino, Takeshi Yagi. Germline mutation rates and the long-term phenotypic effects of mutation accumulation in wild-type laboratory mice and mutator mice. Genome Research 25, 1125-1134(2015). doi:10.1101/gr.186148.114
Sae Tanaka, Junko Tanaka, Yoshihiro Miwa, Daiki D. Horikawa, Toshiaki Katayama, Kazuharu Arakawa, Atsushi Toyoda, Takeo Kubo, Takekazu Kunieda. Novel Mitochondria-Targeted Heat-Soluble Proteins Identified in the Anhydrobiotic Tardigrade Improve Osmotic Tolerance of Human Cells. PLOS ONE 10, e0118272(2015). doi:10.1371/journal.pone.0118272
Matthew R. Shenton, Hajime Ohyanagi, Zi-Xuan Wang, Atsushi Toyoda, Asao Fujiyama, Toshifumi Nagata, Qi Feng, Bin Han, Nori Kurata. Rapid turnover of antimicrobial-type cysteine-rich protein genes in closely related Oryza genomes. Molecular Genetics and Genomics 290, 1753-1770(2015). doi:10.1007/s00438-015-1028-4
Kengo Sato, Yoko Kuroki, Wakako Kumita, Asao Fujiyama, Atsushi Toyoda, Jun Kawai, Atsushi Iriki, Erika Sasaki, Hideyuki Okano, Yasubumi Sakakibara. Resequencing of the common marmoset genome improves genome assemblies and gene-coding sequence analysis. Scientific Reports 5, (2015). doi:10.1038/srep16894
Marinela Perpelescu, Tetsuya Hori, Atsushi Toyoda, Sadahiko Misu, Norikazu Monma, Kazuho Ikeo, Chikashi Obuse, Asao Fujiyama, Tatsuo Fukagawa. HJURP is involved in the expansion of centromeric chromatin. Molecular Biology of the Cell 26, 2742-2754(2015). doi:10.1091/mbc.E15-02-0094
Lizabeth A. Perkins, Laura Holderbaum, Rong Tao, Yanhui Hu, Richelle Sopko, Kim McCall, Donghui Yang-Zhou, Ian Flockhart, Richard Binari, Hye-Seok Shim, Audrey Miller, Amy Housden, Marianna Foos, Sakara Randkelv, Colleen Kelley, Pema Namgyal, Christians Villalta, Lu-Ping Liu, Xia Jiang, Qiao Huan-Huan, Xia Wang, Asao Fujiyama, Atsushi Toyoda, Kathleen Ayers, Allison Blum, Benjamin Czech, Ralph Neumuller, Dong Yan, Amanda Cavallaro, Karen Hibbard, Don Hall, Lynn Cooley, Gregory J. Hannon, Ruth Lehmann, Annette Parks, Stephanie E. Mohr, Ryu Ueda, Shu Kondo, Jian-Quan Ni, Norbert Perrimon. The Transgenic RNAi Project at Harvard Medical School: Resources and Validation. Genetics 201, 843-852(2015). doi:10.1534/genetics.115.180208
Shosei Kubota, Takaya Iwasaki, Kousuke Hanada, Atsushi J. Nagano, Asao Fujiyama, Atsushi Toyoda, Sumio Sugano, Yutaka Suzuki, Kouki Hikosaka, Motomi Ito, Shin-Ichi Morinaga. A Genome Scan for Genes Underlying Microgeographic-Scale Local Adaptation in a Wild Arabidopsis Species. PLOS Genetics 11, e1005361(2015). doi:10.1371/journal.pgen.1005361
Shosei Kubota, Takaya Iwasaki, Kousuke Hanada, Atsushi J. Nagano, Asao Fujiyama, Atsushi Toyoda, Sumio Sugano, Yutaka Suzuki, Kouki Hikosaka, Motomi Ito, Shin-Ichi Morinaga. Correction: A Genome Scan for Genes Underlying Microgeographic-Scale Local Adaptation in a Wild Arabidopsis Species. PLOS Genetics 11, e1005488(2015). doi:10.1371/journal.pgen.1005488
Hiromi Kato, Natsumi Ogawa, Yoshiyuki Ohtsubo, Kenshiro Oshima, Atsushi Toyoda, Hiroshi Mori, Yuji Nagata, Ken Kurokawa, Masahira Hattori, Asao Fujiyama, Masataka Tsuda. Complete Genome Sequence of a Phenanthrene Degrader, Mycobacterium sp. Strain EPa45 (NBRC 110737), Isolated from a Phenanthrene-Degrading Consortium. Genome Announcements 3, (2015). doi:10.1128/genomeA.00782-15
Hiromi Kato, Hiroshi Mori, Fumito Maruyama, Atsushi Toyoda, Kenshiro Oshima, Ryo Endo, Genki Fuchu, Masatoshi Miyakoshi, Ayumi Dozono, Yoshiyuki Ohtsubo, Yuji Nagata, Masahira Hattori, Asao Fujiyama, Ken Kurokawa, Masataka Tsuda. Time-series metagenomic analysis reveals robustness of soil microbiome against chemical disturbance. DNA Research 22, 413-424(2015). doi:10.1093/dnares/dsv023
Minako Izutsu, Atsushi Toyoda, Asao Fujiyama, Kiyokazu Agata, Naoyuki Fuse. Dynamics of Dark-Fly Genome Under Environmental Selections. G3: Genes, Genomes, Genetics 6, 365-376(2015). doi:10.1534/g3.115.023549
Tasuku Ito, Yoshiaki Tarutani, Taiko Kim To, Mohamed Kassam, Evelyne Duvernois-Berthet, Sandra Cortijo, Kazuya Takashima, Hidetoshi Saze, Atsushi Toyoda, Asao Fujiyama, Vincent Colot, Tetsuji Kakutani. Genome-Wide Negative Feedback Drives Transgenerational DNA Methylation Dynamics in Arabidopsis. PLOS Genetics 11, e1005154(2015). doi:10.1371/journal.pgen.1005154
Yang An, Atsushi Toyoda, Chen Zhao, Asao Fujiyama, Kiyokazu Agata. A Colony Multiplex Quantitative PCR-Based 3S3DBC Method and Variations of It for Screening DNA Libraries. PLOS ONE 10, e0116997(2015). doi:10.1371/journal.pone.0116997
- 2014年 -
Kohta Yoshida, Takashi Makino, Katsushi Yamaguchi, Shuji Shigenobu, Mitsuyasu Hasebe, Masakado Kawata, Manabu Kume, Seiichi Mori, Catherine L. Peichel, Atsushi Toyoda, Asao Fujiyama, Jun Kitano. Sex Chromosome Turnover Contributes to Genomic Divergence between Incipient Stickleback Species. PLoS Genetics 10, e1004223(2014). doi:10.1371/journal.pgen.1004223
Yusuke Takehana, Masaru Matsuda, Taijun Myosho, Maximiliano L. Suster, Koichi Kawakami, Tadasu Shin-I, Yuji Kohara, Yoko Kuroki, Atsushi Toyoda, Asao Fujiyama, Satoshi Hamaguchi, Mitsuru Sakaizumi, Kiyoshi Naruse. Co-option of Sox3 as the male-determining factor on the Y chromosome in the fish Oryzias dancena. Nature Communications 5, (2014). doi:10.1038/ncomms5157
Mikhail Spivakov, Thomas O. Auer, Ravindra Peravali, Ian Dunham, Dirk Dolle, Asao Fujiyama, Atsushi Toyoda, Tomoyuki Aizu, Yohei Minakuchi, Felix Loosli, Kiyoshi Naruse, Ewan Birney, Joachim Wittbrodt. Genomic and Phenotypic Characterization of a Wild Medaka Population: Towards the Establishment of an Isogenic Population Genetic Resource in Fish. G3: Genes, Genomes, Genetics 4, 433-445(2014). doi:10.1534/g3.113.008722
Kohei Shimono, Kazuto Fujishima, Takafumi Nomura, Masayoshi Ohashi, Tadao Usui, Mineko Kengaku, Atsushi Toyoda, Tadashi Uemura. An evolutionarily conserved protein CHORD regulates scaling of dendritic arbors with body size. Scientific Reports 4, (2014). doi:10.1038/srep04415
M. Sakurai, H. Ueda, T. Yano, S. Okada, H. Terajima, T. Mitsuyama, A. Toyoda, A. Fujiyama, H. Kawabata, T. Suzuki. A biochemical landscape of A-to-I RNA editing in the human brain transcriptome. Genome Research 24, 522-534(2014). doi:10.1101/gr.162537.113
Masaaki Kobayashi, Hideki Nagasaki, Virginie Garcia, Daniel Just, Cécile Bres, Jean-Philippe Mauxion, Marie-Christine Le Paslier, Dominique Brunel, Kunihiro Suda, Yohei Minakuchi, Atsushi Toyoda, Asao Fujiyama, Hiromi Toyoshima, Takayuki Suzuki, Kaori Igarashi, Christophe Rothan, Eli Kaminuma, Yasukazu Nakamura, Kentaro Yano, Koh Aoki. Genome-Wide Analysis of Intraspecific DNA Polymorphism in ‘Micro-Tom’, a Model Cultivar of Tomato (Solanum lycopersicum). Plant and Cell Physiology 55, 445-454(2014). doi:10.1093/pcp/pct181
Hiroko Kawai-Toyooka, Toshiyuki Mori, Takashi Hamaji, Masahiro Suzuki, Bradley J. S. C. Olson, Tomohiro Uemura, Takashi Ueda, Akihiko Nakano, Atsushi Toyoda, Asao Fujiyama, Hisayoshi Nozaki. Sex-Specific Posttranslational Regulation of the Gamete Fusogen GCS1 in the Isogamous Volvocine Alga Gonium pectorale. Eukaryotic Cell 13, 648-656(2014). doi:10.1128/EC.00330-13
Mikihiko Kawai, Taiki Futagami, Atsushi Toyoda, Yoshihiro Takaki, Shinro Nishi, Sayaka Hori, Wataru Arai, Taishi Tsubouchi, Yuki Morono, Ikuo Uchiyama, Takehiko Ito, Asao Fujiyama, Fumio Inagaki, Hideto Takami. High frequency of phylogenetically diverse reductive dehalogenase-homologous genes in deep subseafloor sedimentary metagenomes. Frontiers in Microbiology 5, (2014). doi:10.3389/fmicb.2014.00080
Mayumi Kamada, Sumitaka Hase, Kengo Sato, Atsushi Toyoda, Asao Fujiyama, Yasubumi Sakakibara. Whole Genome Complete Resequencing of Bacillus subtilis Natto by Combining Long Reads with High-Quality Short Reads. PLoS ONE 9, e109999(2014). doi:10.1371/journal.pone.0109999
Rei Kajitani, Kouta Toshimoto, Hideki Noguchi, Atsushi Toyoda, Yoshitoshi Ogura, Miki Okuno, Mitsuru Yabana, Masayuki Harada, Eiji Nagayasu, Haruhiko Maruyama, Yuji Kohara, Asao Fujiyama, Tetsuya Hayashi, Takehiko Itoh. Efficient de novo assembly of highly heterozygous genomes from whole-genome shotgun short reads. Genome Research 24, 1384-1395(2014). doi:10.1101/gr.170720.113
Tetsuya Hori, Wei-Hao Shang, Atsushi Toyoda, Sadahiko Misu, Norikazu Monma, Kazuho Ikeo, Oscar Molina, Giulia Vargiu, Asao Fujiyama, Hiroshi Kimura, William C. Earnshaw, Tatsuo Fukagawa. Histone H4 Lys 20 Monomethylation of the CENP-A Nucleosome Is Essential for Kinetochore Assembly. Developmental Cell 29, 740-749(2014). doi:10.1016/j.devcel.2014.05.001
- 2013年 -
H. Mori, F. Maruyama, H. Kato, A. Toyoda, A. Dozono, Y. Ohtsubo, Y. Nagata, A. Fujiyama, M. Tsuda, K. Kurokawa. Design and Experimental Application of a Novel Non-Degenerate Universal Primer Set that Amplifies Prokaryotic 16S rRNA Genes with a Low Possibility to Amplify Eukaryotic rRNA Genes. DNA Research 21, 217-227(2013). doi:10.1093/dnares/dst052
Yuji Takahashi, Yoko Fukuda, Jun Yoshimura, Atsushi Toyoda, Kari Kurppa, Hiroyoko Moritoyo, Veronique V. Belzil, Patrick A. Dion, Koichiro Higasa, Koichiro Doi, Hiroyuki Ishiura, Jun Mitsui, Hidetoshi Date, Budrul Ahsan, Takashi Matsukawa, Yaeko Ichikawa, Takashi Moritoyo, Mayumi Ikoma, Tsukasa Hashimoto, Fumiharu Kimura, Shigeo Murayama, Osamu Onodera, Masatoyo Nishizawa, Mari Yoshida, Naoki Atsuta, Gen Sobue, Jennifer A. Fifita, Kelly L. Williams, Ian P. Blair, Garth A. Nicholson, Paloma Gonzalez-Perez, Robert H. Brown, Masahiro Nomoto, Klaus Elenius, Guy A. Rouleau, Asao Fujiyama, Shinichi Morishita, Jun Goto, Shoji Tsuji. ERBB4 Mutations that Disrupt the Neuregulin-ErbB4 Pathway Cause Amyotrophic Lateral Sclerosis Type 19. The American Journal of Human Genetics 93, 900-905(2013). doi:10.1016/j.ajhg.2013.09.008
T. Takada, T. Ebata, H. Noguchi, T. M. Keane, D. J. Adams, T. Narita, T. Shin-I, H. Fujisawa, A. Toyoda, K. Abe, Y. Obata, Y. Sakaki, K. Moriwaki, A. Fujiyama, Y. Kohara, T. Shiroishi. The ancestor of extant Japanese fancy mice contributed to the mosaic genomes of classical inbred strains. Genome Research 23, 1329-1338(2013). doi:10.1101/gr.156497.113
Eiichi Shoguchi, Chuya Shinzato, Takeshi Kawashima, Fuki Gyoja, Sutada Mungpakdee, Ryo Koyanagi, Takeshi Takeuchi, Kanako Hisata, Makiko Tanaka, Mayuki Fujiwara, Mayuko Hamada, Azadeh Seidi, Manabu Fujie, Takeshi Usami, Hiroki Goto, Shinichi Yamasaki, Nana Arakaki, Yutaka Suzuki, Sumio Sugano, Atsushi Toyoda, Yoko Kuroki, Asao Fujiyama, Mónica Medina, Mary Alice Coffroth, Debashish Bhattacharya, Nori Satoh. Draft Assembly of the Symbiodinium minutum Nuclear Genome Reveals Dinoflagellate Gene Structure. Current Biology 23, 1399-1408(2013). doi:10.1016/j.cub.2013.05.062
Daisuke Shiomi, Atsushi Toyoda, Tomoyuki Aizu, Fumio Ejima, Asao Fujiyama, Tadasu Shini, Yuji Kohara, Hironori Niki. Mutations in cell elongation genesmreB,mrdAandmrdBsuppress the shape defect of RodZ-deficient cells. Molecular Microbiology 87, 1029-1044(2013). doi:10.1111/mmi.12148
Wei-Hao Shang, Tetsuya Hori, Nuno M.C. Martins, Atsushi Toyoda, Sadahiko Misu, Norikazu Monma, Ichiro Hiratani, Kazuhiro Maeshima, Kazuho Ikeo, Asao Fujiyama, Hiroshi Kimura, William C. Earnshaw, Tatsuo Fukagawa. Chromosome Engineering Allows the Efficient Isolation of Vertebrate Neocentromeres. Developmental Cell 24, 635-648(2013). doi:10.1016/j.devcel.2013.02.009
Masato Nikaido, Hikoyu Suzuki, Atsushi Toyoda, Asao Fujiyama, Kimiko Hagino-Yamagishi, Thomas D. Kocher, Karen Carleton, Norihiro Okada. Lineage-Specific Expansion of Vomeronasal Type 2 Receptor-Like (OlfC) Genes in Cichlids May Contribute to Diversification of Amino Acid Detection Systems. Genome Biology and Evolution 5, 711-722(2013). doi:10.1093/gbe/evt041
M. Nikaido, H. Noguchi, H. Nishihara, A. Toyoda, Y. Suzuki, R. Kajitani, H. Suzuki, M. Okuno, M. Aibara, B. P. Ngatunga, S. I. Mzighani, H. W. J. Kalombo, K. W. A. Masengi, J. Tuda, S. Nogami, R. Maeda, M. Iwata, Y. Abe, K. Fujimura, M. Okabe, T. Amano, A. Maeno, T. Shiroishi, T. Itoh, S. Sugano, Y. Kohara, A. Fujiyama, N. Okada. Coelacanth genomes reveal signatures for evolutionary transition from water to land. Genome Research 23, 1740-1748(2013). doi:10.1101/gr.158105.113
Miyake Kunio, Chunshu Yang, Yohei Minakuchi, Kenta Ohori, Masaki Soutome, Takae Hirasawa, Yasuhiro Kazuki, Noboru Adachi, Seiko Suzuki, Masayuki Itoh, Yu-ichi Goto, Tomoko Andoh, Hiroshi Kurosawa, Wado Akamatsu, Manabu Ohyama, Hideyuki Okano, Mitsuo Oshimura, Masayuki Sasaki, Atsushi Toyoda, Takeo Kubota. Comparison of Genomic and Epigenomic Expression in Monozygotic Twins Discordant for Rett Syndrome. PLoS ONE 8, e66729(2013). doi:10.1371/journal.pone.0066729
Daiki D. Horikawa, John Cumbers, Iori Sakakibara, Dana Rogoff, Stefan Leuko, Raechel Harnoto, Kazuharu Arakawa, Toshiaki Katayama, Takekazu Kunieda, Atsushi Toyoda, Asao Fujiyama, Lynn J. Rothschild. Analysis of DNA Repair and Protection in the Tardigrade Ramazzottius varieornatus and Hypsibius dujardini after Exposure to UVC Radiation. PLoS ONE 8, e64793(2013). doi:10.1371/journal.pone.0064793
Takashi Hamaji, David R. Smith, Hideki Noguchi, Atsushi Toyoda, Masahiro Suzuki, Hiroko Kawai-Toyooka, Asao Fujiyama, Ichiro Nishii, Tara Marriage, Bradley J. S. C. Olson, Hisayoshi Nozaki. Mitochondrial and Plastid Genomes of the Colonial Green Alga Gonium pectorale Give Insights into the Origins of Organelle DNA Architecture within the Volvocales. PLoS ONE 8, e57177(2013). doi:10.1371/journal.pone.0057177
Yu Fu, Akira Kawabe, Mathilde Etcheverry, Tasuku Ito, Atsushi Toyoda, Asao Fujiyama, Vincent Colot, Yoshiaki Tarutani, Tetsuji Kakutani. Mobilization of a plant transposon by expression of the transposon-encoded anti-silencing factor. The EMBO Journal 32, 2407-2417(2013). doi:10.1038/emboj.2013.169
- 2012年 -
Y. Yamamoto, T. Watanabe, Y. Hoki, K. Shirane, Y. Li, K. Ichiiyanagi, S. Kuramochi-Miyagawa, A. Toyoda, A. Fujiyama, M. Oginuma, H. Suzuki, T. Sado, T. Nakano, H. Sasaki. Targeted gene silencing in mouse germ cells by insertion of a homologous DNA into a piRNA generating locus. Genome Research 23, 292-299(2012). doi:10.1101/gr.137224.112
Ayami Yamaguchi, Sae Tanaka, Shiho Yamaguchi, Hirokazu Kuwahara, Chizuko Takamura, Shinobu Imajoh-Ohmi, Daiki D. Horikawa, Atsushi Toyoda, Toshiaki Katayama, Kazuharu Arakawa, Asao Fujiyama, Takeo Kubo, Takekazu Kunieda. Two Novel Heat-Soluble Protein Families Abundantly Expressed in an Anhydrobiotic Tardigrade. PLoS ONE 7, e44209(2012). doi:10.1371/journal.pone.0044209
S. Tsukahara, A. Kawabe, A. Kobayashi, T. Ito, T. Aizu, T. Shin-i, A. Toyoda, A. Fujiyama, Y. Tarutani, T. Kakutani. Centromere-targeted de novo integrations of an LTR retrotransposon of Arabidopsis lyrata. Genes & Development 26, 705-713(2012). doi:10.1101/gad.183871.111
Hideto Takami, Hideki Noguchi, Yoshihiro Takaki, Ikuo Uchiyama, Atsushi Toyoda, Shinro Nishi, Gab-Joo Chee, Wataru Arai, Takuro Nunoura, Takehiko Itoh, Masahira Hattori, Ken Takai. A Deeply Branching Thermophilic Bacterium with an Ancient Acetyl-CoA Pathway Dominates a Subsurface Ecosystem. PLoS ONE 7, e30559(2012). doi:10.1371/journal.pone.0030559
Yuuta Moriyama, Toru Kawanishi, Ryohei Nakamura, Tatsuya Tsukahara, Kenta Sumiyama, Maximiliano L. Suster, Koichi Kawakami, Atsushi Toyoda, Asao Fujiyama, Yuuri Yasuoka, Yusuke Nagao, Etsuko Sawatari, Atsushi Shimizu, Yuko Wakamatsu, Masahiko Hibi, Masanori Taira, Masataka Okabe, Kiyoshi Naruse, Hisashi Hashimoto, Atsuko Shimada, Hiroyuki Takeda. The Medaka zic1/zic4 Mutant Provides Molecular Insights into Teleost Caudal Fin Evolution. Current Biology 22, 601-607(2012). doi:10.1016/j.cub.2012.01.063
Sumiyo Morita, Ryou-u Takahashi, Riu Yamashita, Atsushi Toyoda, Takuro Horii, Mika Kimura, Asao Fujiyama, Kenta Nakai, Shoji Tajima, Ryo Matoba, Takahiro Ochiya, Izuho Hatada. Genome-Wide Analysis of DNA Methylation and Expression of MicroRNAs in Breast Cancer Cells. International Journal of Molecular Sciences 13, 8259-8272(2012). doi:10.3390/ijms13078259
R. N. Kim, D.-S. Kim, S.-H. Choi, B.-H. Yoon, A. Kang, S.-H. Nam, D.-W. Kim, J.-J. Kim, J.-H. Ha, A. Toyoda, A. Fujiyama, A. Kim, M.-Y. Kim, K.-H. Park, K. S. Lee, H.-S. Park. Genome Analysis of the Domestic Dog (Korean Jindo) by Massively Parallel Sequencing. DNA Research 19, 275-288(2012). doi:10.1093/dnares/dss011
Kagoshima, H., Kito, K., Aizu, T., Shin-I, T., Kanda, H., Kobayashi, S., Toyoda, A., Fujiyama, A., Kohara, Y., Convey, P. and Niki, H. Multi-decadal survival of an Antarctic nematode, Plectus murrayi, in a -20 degrees C stored moss sample. CryoLetters 33, 280-288(2012). PMID: 22987239
Minako Izutsu, Jun Zhou, Yuzo Sugiyama, Osamu Nishimura, Tomoyuki Aizu, Atsushi Toyoda, Asao Fujiyama, Kiyokazu Agata, Naoyuki Fuse. Genome Features of “Dark-Fly”, a Drosophila Line Reared Long-Term in a Dark Environment. PLoS ONE 7, e33288(2012). doi:10.1371/journal.pone.0033288
Xuehui Huang, Nori Kurata, Xinghua Wei, Zi-Xuan Wang, Ahong Wang, Qiang Zhao, Yan Zhao, Kunyan Liu, Hengyun Lu, Wenjun Li, Yunli Guo, Yiqi Lu, Congcong Zhou, Danlin Fan, Qijun Weng, Chuanrang Zhu, Tao Huang, Lei Zhang, Yongchun Wang, Lei Feng, Hiroyasu Furuumi, Takahiko Kubo, Toshie Miyabayashi, Xiaoping Yuan, Qun Xu, Guojun Dong, Qilin Zhan, Canyang Li, Asao Fujiyama, Atsushi Toyoda, Tingting Lu, Qi Feng, Qian Qian, Jiayang Li, Bin Han. A map of rice genome variation reveals the origin of cultivated rice. Nature 490, 497-501(2012). doi:10.1038/nature11532
Hideki Hirakawa, Suguru Tsuchimoto, Hiroe Sakai, Shinobu Nakayama, Tsunakazu Fujishiro, Yoshie Kishida, Mitsuyo Kohara, Akiko Watanabe, Manabu Yamada, Tomoyuki Aizu, Atsushi Toyoda, Asao Fujiyama, Satoshi Tabata, Kiichi Fukui, Shusei Sato. Upgraded genomic information of Jatropha curcas L.. Plant Biotechnology 29, 123-130(2012). doi:10.5511/plantbiotechnology.12.0515a
- 2011年 -
V. J. Murtagh, D. O'Meally, N. Sankovic, M. L. Delbridge, Y. Kuroki, J. L. Boore, A. Toyoda, K. S. Jordan, A. J. Pask, M. B. Renfree, A. Fujiyama, J. A. M. Graves, P. D. Waters. Evolutionary history of novel genes on the tammar wallaby Y chromosome: Implications for sex chromosome evolution. Genome Research 22, 498-507(2011). doi:10.1101/gr.120790.111
T. Watanabe, S.-i. Tomizawa, K. Mitsuya, Y. Totoki, Y. Yamamoto, S. Kuramochi-Miyagawa, N. Iida, Y. Hoki, P. J. Murphy, A. Toyoda, K. Gotoh, H. Hiura, T. Arima, A. Fujiyama, T. Sado, T. Shibata, T. Nakano, H. Lin, K. Ichiyanagi, P. D. Soloway, H. Sasaki. Role for piRNAs and Noncoding RNA in de Novo DNA Methylation of the Imprinted Mouse Rasgrf1 Locus. Science 332, 848-852(2011). doi:10.1126/science.1203919
Toshiaki Watanabe, Shinichiro Chuma, Yasuhiro Yamamoto, Satomi Kuramochi-Miyagawa, Yasushi Totoki, Atsushi Toyoda, Yuko Hoki, Asao Fujiyama, Tatsuhiro Shibata, Takashi Sado, Toshiaki Noce, Toru Nakano, Norio Nakatsuji, Haifan Lin, Hiroyuki Sasaki. MITOPLD Is a Mitochondrial Protein Essential for Nuage Formation and piRNA Biogenesis in the Mouse Germline. Developmental Cell 20, 364-375(2011). doi:10.1016/j.devcel.2011.01.005
Marilyn B Renfree, Anthony T Papenfuss, Janine E Deakin, James Lindsay, Thomas Heider, Katherine Belov, Willem Rens, Paul D Waters, Elizabeth A Pharo, Geoff Shaw, Emily SW Wong, Christophe M Lefèvre, Kevin R Nicholas, Yoko Kuroki, Matthew J Wakefield, Kyall R Zenger, Chenwei Wang, Malcolm Ferguson-Smith, Frank W Nicholas, Danielle Hickford, Hongshi Yu, Kirsty R Short, Hannah V Siddle, Stephen R Frankenberg, Keng Chew, Brandon R Menzies, Jessica M Stringer, Shunsuke Suzuki, Timothy A Hore, Margaret L Delbridge, Amir Mohammadi, Nanette Y Schneider, Yanqiu Hu, William O'Hara, Shafagh Al Nadaf, Chen Wu, Zhi-Ping Feng, Benjamin G Cocks, Jianghui Wang, Paul Flicek, Stephen MJ Searle, Susan Fairley, Kathryn Beal, Javier Herrero, Dawn M Carone, Yutaka Suzuki, Sumio Sugano, Atsushi Toyoda, Yoshiyuki Sakaki, Shinji Kondo, Yuichiro Nishida, Shoji Tatsumoto, Ion Mandiou, Arthur Hsu, Kaighin A McColl, Benjamin Lansdell, George Weinstock, Elizabeth Kuczek, Annette McGrath, Peter Wilson, Artem Men, Mehlika Hazar-Rethinam, Allison Hall, John Davis, David Wood, Sarah Williams, Yogi Sundaravadanam, Donna M Muzny, Shalini N Jhangiani, Lora R Lewis, Margaret B Morgan, Geoffrey O Okwuonu, San Ruiz, Jireh Santibanez, Lynne Nazareth, Andrew Cree, Gerald Fowler, Christie L Kovar, Huyen H Dinh, Vandita Joshi, Chyn Jing, Fremiet Lara, Rebecca Thornton, Lei Chen, Jixin Deng, Yue Liu, Joshua Y Shen, Xing-Zhi Song, Janette Edson, Carmen Troon, Daniel Thomas, Amber Stephens, Lankesha Yapa, Tanya Levchenko, Richard A Gibbs, Desmond W Cooper, Terence P Speed, Asao Fujiyama, Jennifer A M Graves, Rachel J O'Neill, Andrew J Pask, Susan M Forrest, Kim C Worley. Genome sequence of an Australian kangaroo, Macropus eugenii, provides insight into the evolution of mammalian reproduction and development. Genome Biology 12, R81(2011). doi:10.1186/gb-2011-12-8-r81
R. Ono, Y. Kuroki, M. Naruse, M. Ishii, S. Iwasaki, A. Toyoda, A. Fujiyama, G. Shaw, M. B. Renfree, T. Kaneko-Ishino, F. Ishino. Identification of tammar wallaby SIRH12, derived from a marsupial-specific retrotransposition event. DNA Research 18, 211-219(2011). doi:10.1093/dnares/dsr012
Wanyang Liu, Daisuke Morito, Seiji Takashima, Yohei Mineharu, Hatasu Kobayashi, Toshiaki Hitomi, Hirokuni Hashikata, Norio Matsuura, Satoru Yamazaki, Atsushi Toyoda, Ken-ichiro Kikuta, Yasushi Takagi, Kouji H. Harada, Asao Fujiyama, Roman Herzig, Boris Krischek, Liping Zou, Jeong Eun Kim, Masafumi Kitakaze, Susumu Miyamoto, Kazuhiro Nagata, Nobuo Hashimoto, Akio Koizumi. Identification of RNF213 as a Susceptibility Gene for Moyamoya Disease and Its Possible Role in Vascular Development. PLoS ONE 6, e22542(2011). doi:10.1371/journal.pone.0022542
S. Sato, H. Hirakawa, S. Isobe, E. Fukai, A. Watanabe, M. Kato, K. Kawashima, C. Minami, A. Muraki, N. Nakazaki, C. Takahashi, S. Nakayama, Y. Kishida, M. Kohara, M. Yamada, H. Tsuruoka, S. Sasamoto, S. Tabata, T. Aizu, A. Toyoda, T. Shin-i, Y. Minakuchi, Y. Kohara, A. Fujiyama, S. Tsuchimoto, S. Kajiyama, E. Makigano, N. Ohmido, N. Shibagaki, J. A. Cartagena, N. Wada, T. Kohinata, A. Atefeh, S. Yuasa, S. Matsunaga, K. Fukui. Sequence Analysis of the Genome of an Oil-Bearing Tree, Jatropha curcas L.. DNA Research 18, 65-76(2011). doi:10.1093/dnares/dsq030
| 項目 | 2024年度 | 2023年度 | ||
|---|---|---|---|---|
| 総塩基数 | % | 総塩基数 | % | |
| ゲノム解析 | 27,663,142,963,000 | 57.2 | 23,708,774,949,900 | 48.3 |
| 高次構造解析と修飾解析 | 14,012,117,387,580 | 29.0 | 17,553,845,677,600 | 35.8 |
| トランスクリプトーム解析 | 4,499,587,229,550 | 9.3 | 6,032,704,123,390 | 12.3 |
| アンプリコン解析 | 253,447,851,400 | 0.5 | 626,558,241,000 | 1.3 |
| シングルセル解析 | 1,961,238,670,482 | 4.1 | 1,115,576,237,200 | 2.3 |
| エキソーム解析 | 0 | 0.0 | 0 | 0.0 |
| 合計 | 48,389,534,102,012 | 49,037,459,229,090 | ||
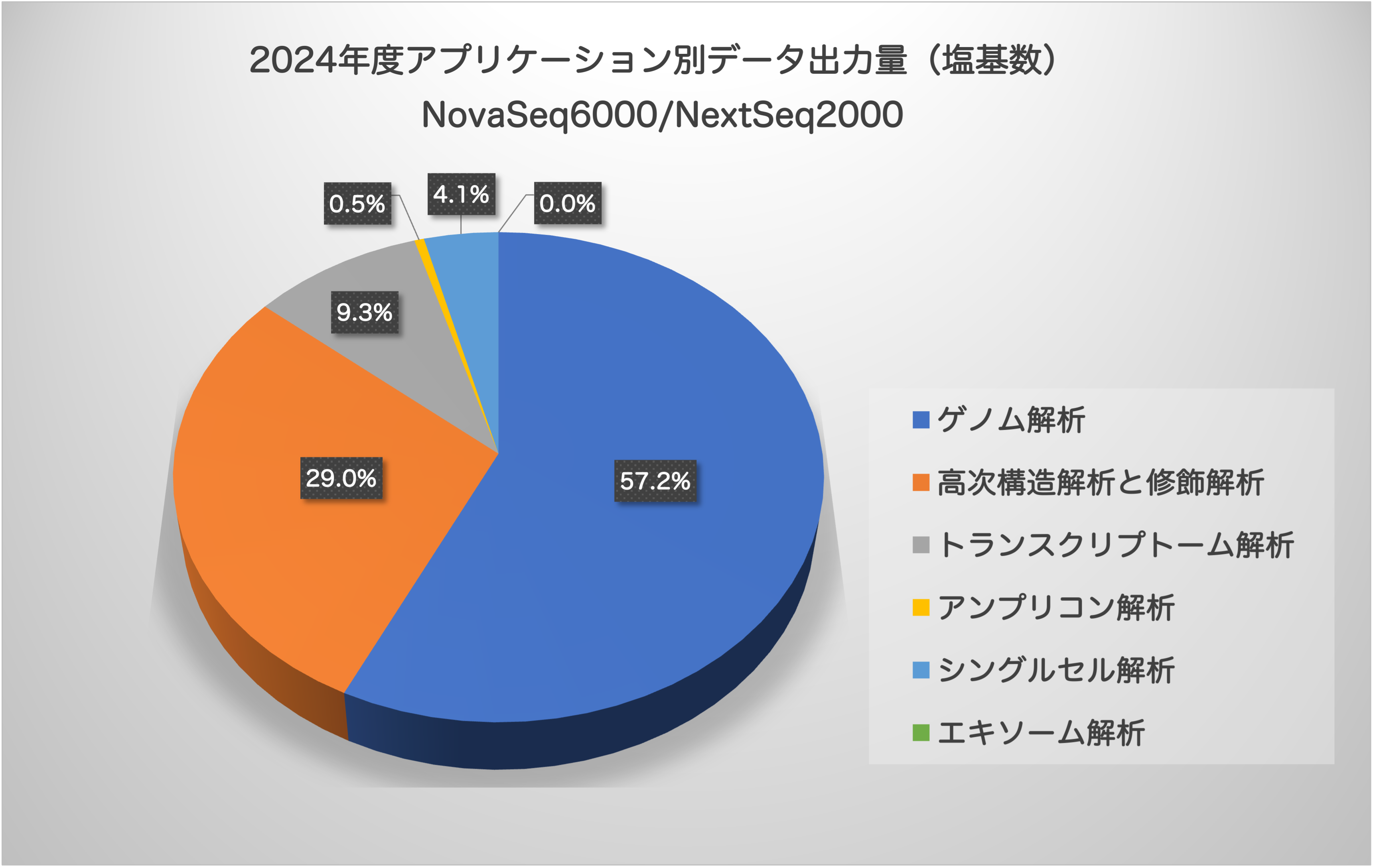
| 項目 | 2024年度 | 2023年度 | ||
|---|---|---|---|---|
| 総塩基数 | % | 総塩基数 | % | |
| ゲノム解析 | 125,994,009,229,779 | 70.1 | 65,701,146,323,851 | 75.7 |
| その他(Iso-Seq解析など) | 53,643,623,580,906 | 29.9 | 21,056,017,367,421 | 24.3 |
| 合計 | 179,637,632,810,685 | 86,757,163,691,272 | ||
※2024年度はRevioとSequel IIeを利用



